Abstract
Bacteroides fragilis is a minor component of the intestinal microbiota and the most frequently isolated from intra-abdominal infections and bacteremia. Previously, our group has shown that molecules involved in laminin-1 (LMN-1) recognition were present in outer membrane protein extracts of B. fragilis MC2 strain. One of these proteins was identified and showed 98% similarity to a putative B. fragilis plasminogen-binding protein precursor, deposited in the public database. Thus, the objective of this work was to overexpress and further characterize this novel adhesin. The ability of B. fragilis MC2 strain and purified protein to convert plasminogen into plasmin was tested. Our results showed that B. fragilis strain MC2 strain adhered to both LMN-1 and plasminogen and this adhesion was inhibited by either LMN-1 or plasminogen. Regarding the plasminogen activation activity, both the whole bacterial cell and the purified protein converted plasminogen into plasmin similar to streptokinase used as a positive control. Bacterial receptors that recognize plasminogen bind to it and enhance its activation, transforming a nonproteolytic bacterium into a proteolytic one. We present in vitro evidence for a pathogenic function of the plasminogen receptor in promoting adherence to laminin and also the formation of plasmin by B. fragilis.
Keywords: Bacteroides fragilis, plasminogen, fibrinolytic system
Introduction
Bacteroides fragilis is a Gram-negative strictly anaerobic bacterium that represents about 1% of the total Bacteroides found in the human large intestinal tract (Finegold, 1995). Although it is a minor component of the intestinal microbiota, B. fragilis is the most frequently isolated bacterium from clinical infections, especially from patients with intra-abdominal infections (Gibson et al., 1998) and bacteremia (Brook & Frasier, 2000). It is also isolated from brain abscesses, deep wounds and female genital tract infections (Shah, 1999). Several virulence factors have been described for this microorganism, such as proteases (Patrick et al., 1996), enterotoxin (Wu et al., 1998) and lipopolysaccharide (Pumbwe et al., 2006), but their roles in pathogenicity are still not well elucidated. The most-studied virulence factor is the capsular polysaccharide complex (CPC). Bacteroides fragilis CPC can not only modulate its surface antigenicity by expressing at least eight distinct polysaccharides (Krinos et al., 2001), but are also directly involved in abscess formation in in vivo experimental infection models (Gibson et al., 1998).
Recently, a putative plasminogen-binding protein, Bfp60, was identified in B. fragilis BE1 and it was suggested that this property might contribute to the virulence of this bacterium (Sijbrandi et al., 2005). Plasminogen is a 92-kDa glycoprotein that is converted to an active serine protease plasmin either by the urokinase-type plasminogen activator or by the tissue-type plasminogen activator (tPA), in the fibrinolytic system (Sun, 2006). Indiscriminate activation of plasmin can cause significant tissue damage, and for this reason, the recruitment and activation of plasminogen must be well controlled (Lahteenmaki et al., 2001). Many invasive human bacterial pathogens, such as Neisseria meningitidis (Ullberg et al., 1992), Salmonella typhimurium (Lähteenmäki et al., 1995), Staphylococcus aureus (Mökänen et al., 2002) and Helicobacter pylori (Jönsson et al., 2004), interfere with the mammalian plasminogen/plasmin system by expressing plasminogen receptors or activators (Lähteenmäki et al., 2001). The bacterial receptors that recognize plasminogen bind to it and enhance its activation by tPA on the bacterial surface, transforming a nonproteolytic bacterium into a proteolytic one with the help of a host-derived proteolytic system. It has been speculated that because the metastatic tumor cells utilize plasminogen activation to penetrate the basal membrane (BM), the bacteria would function in an analogous way (Boyle & Lottenberg, 1997). In its normal physiological role, plasmin dissolves fibrin clots, but plasmin has also been implicated in the degradation of other substrates, such as the extracellular matrix (ECM) protein components.
Previously, our group has described the ability of two B. fragilis strains, MC2 and 1081, to adhere to laminin-1 (LMN-1) with high affinity (Ferreira et al., 2006). It was also shown that the molecules involved in this recognition were glycoproteins presented in B. fragilis outer membrane protein (OMP) extracts. The aim of this study was to isolate, clone, express and further characterize this putative novel adhesin present on the surface of B. fragilis strains. We show that the LMN-1-binding protein has significant amino acid similarity to the putative binding plasminogen protein precursor, Bfp60. We also demonstrate that this protein binds to both LMN-1 and plasminogen using in vitro adhesion assays and converts plasminogen into plasmin.
Materials and methods
Bacterial strains and growth conditions
The B. fragilis strain MC2, which possesses high affinity to LMN-1 (Ferreira et al., 2006), was used throughout this study. This strain is deposited at the Cell Culture Collection of the Anaerobic Bacteria Laboratory, Instituto Prof. Paulo de Góes – IMPPG, UFRJ, Brazil. Bacteria were grown in prereduced anaerobically sterilized brain heart infusion broth (BHI-PRAS), supplemented with hemin (5 mg mL−1) and l-cystein (0.5 g L−1), for 18 h at 37 °C (Jousiemies Somier et al., 2002).
OMP extraction
The OMPs extracts from B. fragilis strain MC2 strain were obtained according to Bölin et al. (1982). Briefly, the cells were disrupted in a French pressure cell press at 4 °C and intact cells were removed by centrifugation (4000 g) for 10 min. The crude envelope fraction was collected from the supernatant by centrifugation at 100 000 g for 30 min at 4 °C. The pellet was treated twice with a 3% N-lauryl sarcosyl (Sigma) solution and pelleted at 4000 g. Pellets were washed three times with 0.01 M phosphate buffer (pH 7.4) and resuspended in the same buffer. The Bradford dye assay (Sigma) was used to determine the protein concentration. OMP fractions were stored at −20 °C.
Electrophoresis in polyacrylamide gel -- SDS-PAGE
Standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with a discontinuous Bis-acrylamide gel (4% stacking; 12% separating) in a Tris-glycine running buffer (Tris 3 g L−1, glycine 72 g L−1 and SDS 5 g L−1) (Laemmili, 1970). Protein samples were mixed with sample buffer (Tris-HCl, pH 6.8, 10% SDS, 10% glycerol and bromophenol blue 0.05% w/v; Sigma), boiled (100 °C) for 5 min and applied to the gel. Protein standards (Invitrogen) were used as molecular weight markers. All gels were stained with Coomassie brilliant blue R-250.
Affinity column analysis
An affinity column was prepared containing c. 0.5 mL of Affi-gel® 10 (Bio-Rad) (Kern & Schotz, 1987; Lissitsky et al., 2004) and mixed with an LMN-1 (1 mg mL−1) solution in phosphate-buffered saline (PBS) (0.1 M, pH 7.2). After 24 h at 4 °C, 50 mM ethanolamide, pH 7.8, was added and allowed to interact for 1 h. The column was washed three times with 0.1 M PBS, pH 7.2, followed by a wash with 2 M NaCl, pH 7.0. The enriched OMP extracts (1–5 mg mL−1) were passed several times through the column. The column was washed with 0.1 M PBS to remove unbound proteins. Proteins were eluted from the column with a stepwise concentration of NaCl, 0.25, 0.5, 1.0 and 2 M. Fractions were collected and desalted using Centricon concentrators (Amicon®). Samples were analyzed by SDS-PAGE. Pure fractions (proteins eluted with 2 M NaCl) were in-gel trypsin digested and the digested peptides were analyzed by MS/MS spectroscopy for peptide identification at the Proteomics Core Facility at University of North Carolina at Chapel Hill.
Chromosomal extraction
Bacteroides fragilis MC2 cultures (5 mL) were grown overnight (18 h) at 37 °C in BHI-PRAS and 1.5 mL was centrifuged at 5000 g (4 °C) for 2 min, and all DNA modifications and manipulations were carried out using standard methods (Sambrook et al., 1989). The DNA was resuspended in 100 μL of TE containing RNAse A (10 μg mL−1).
Cloning the bfp60 homologue gene
The data obtained from the peptide mass spectroscopy identification matched to a putative plasminogen-binding protein precursor, Bfp60, in B. fragilis BE1 strain (EMBL/GenBank nucleotide database accession number: AJ786264; Sijbrandi et al., 2005). In order to amplify the entire bfp60 homologue gene in the B. fragilis MC2 strain, we used the B. fragilis 638R genome sequence data available at http://www.sanger.ac.uk/Projects/B_fragilis/ to design specific oligonucleotide primers. Primers were designed within corporation of restriction sites as follows: bfp60 forward NdeI (5′TTATACCTCATATGATCAAG3′) and bfp60 reverse XhoI (5′CTCGTAGATTTTATGAGCTCTG3′) to amplify a DNA fragment of 1692 bp containing the entire Bfp60 ORF. The DNA fragment was amplified by PCR using Pfx platinum Taq (Invitrogen) in 50 μL containing 1 × Pfx Taq buffer, 1 mM MgSO4, 0.3 mM dNTP, 0.5 pmol of each primer and 2 μL of DNA (20 μg μL−1). The following cycle program was used: 95 °C for 1 min; 35 cycles at 94 °C for 15 s, 40 °C for 15 s and 72 °C for 2 min; and a final extension at 72 °C for 10 min. The PCR-amplified fragment was purified using the QIAquick gel extraction kit (Qiagen). The amplified gene was sequenced at the Human Genome Research Center (Institute of Biosciences, University of São Paulo) using the MegaBACE™ 1000 and DYEnamic ET Dye Terminator Cycle Sequencing kit (Thermo Sequence™ II DNA polymerase). Nucleotide sequence analysis revealed that the B. fragilis MC2 bfp60 nucleotide region is identical to the bfp60 region in theB. fragilis 638R genome. The amplified fragment was cloned into the NdeI/XhoI sites of the expression vector pET26b+ (Novagen) to constructa recombinant C-terminus 6xHis tag fusion protein, Adhesin-His-tag (Ads-6xHis). The ligation mixture was transformed into competent Escherichia coli DH10B. Restriction digestion with NdeI/XhoI and SmaI/BamHI was used to confirm the plasmid constructs.
Expression and purification of Bfp60
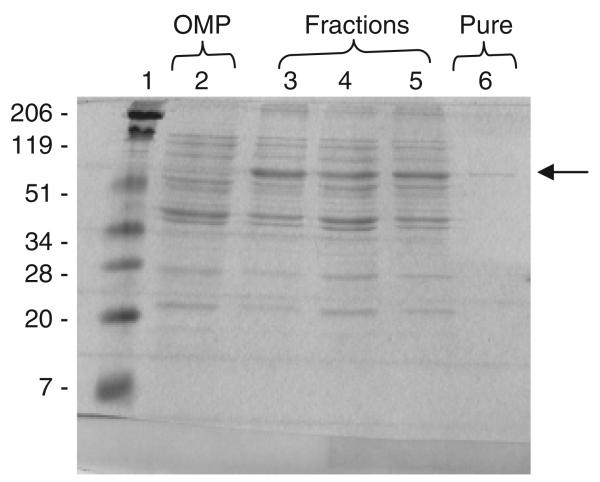
The pET26b+/Ads-6xHis plasmid was transformed into E. coli Rosetta cells for expression of Ads-6xHis. Escherichia coli Rosetta cells were grown in 500 mL of Luria–Bertani medium containing 50 μg mL−1 kanamycin and 25 μg mL−1 chloramphenicol at 37 °C under aeration. After reaching the OD550nm of 0.4–0.5, the cultures were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside and incubated for 4 h under aeration. The cells were harvested at 4000 g at 4 °C and suspended in binding buffer (0.5 M NaCl, 20 mM Tris-HCl and 5 mM imidazole, pH 7.9) containing RNAse (10 μg mL−1), DNAse (10 μg mL−1) and phenylmethylsulfonyl fluoride (1 mM). Bacterial cells were disrupted using a French pressure cell. The lysate was centrifuged at 5000 g for 10 min at 4 °C to remove cell debris. Cell-free supernatants were incubated with 3 mL of a Ni2+-charged resin (Novagen His-Bind® Kits) in a rotary shaker for 1 h at 4 °C. The protein purification was performed according to the manufacturer's instructions at 4 °C. Protein fractions were applied in an SDS-PAGE. The purified protein fractions, hereafter named Bfp60-HIS, were pooled and concentrated in a Pmax Amicon® filter (Fig. 3).
Fig. 3.
Purification of the recombinant Bfp60 protein in SDS-PAGE. Lane 1 – molecular weight standards; lane 2 – whole cell lysates proteins of the Escherichia coli containing the bfp60 gene without the isopropyl β-d-1-thiogalactopyranoside (IPTG) induction; lane 3 – whole cell lysate proteins of E. coli containing the bfp60 gene IPTG-induced; lane 4 – recombinant Bfp60 protein (Bfp60-HP) after purification with Ni-resin. The arrow (→) indicates the size of the recombinant Bfp60-HP protein of 60 kDa.
Adhesion and inhibition assays
Laminin from Engelbreth-Holm-Swarm mouse tumor (LMN-1; Invitrogen) or human plasminogen (Sigma) were used throughout this study. LMN-1 and plasminogen were immobilized onto glass coverslips as described previously (Ferreira et al., 2006) and placed into 24 wells. Briefly, to prepare the plates, 15 μg mL−1 LMN-1 or 10 μg mL−1 plasminogen was resuspended in 0.01 M PBS and immobilized or not onto glass coverslips for 1 h at room temperature. Immediately, LMN-1- or plasminogen-coated coverslips were carefully washed with PBS containing 0.1% bovine serum albumin (BSA) (w/v) in order to remove the unbound LMN-1 or plasminogen, avoiding unspecific association, and blocked with PBS containing 2% BSA (PBSB) for another hour at room temperature. Cover slips coated with 2% BSA were also prepared as control.
For the adhesion assay, 200 μL of a suspension of B. fragilis MC2 cells in 0.1% PBSB (109 CFU mL−1) was added onto the coverslips placed into the 24-well culture plates. For the inhibition assay, the B. fragilis MC2 strain was preincubated with LMN-1 (10 μg mL−1), purified Bfp60-HIS (10 μg mL−1) or plasminogen (10 μg mL−1) in 0.01% PBSB for 1 h at 4 °C before adding the suspension to the coverslips placed in the plate wells. After 1 h of incubation at 37 °C, the coverslips were washed once with 0.01 M PBS, then fixed with methanol for 5 min and further washed with PBS to be stained with 0.1% crystal violet. Three independent experiments were carried out in triplicate and the association of the bacteria (ratio of adherence) to the coverslips was established by counting 10 random fields in each coverslip using an optical microscope (×1000 magnification). Data are presented as mean±SEM (SE of the mean). The Student t-test was used for statistical analysis, and values with P<0.05 were considered significant.
Plasminogen activation assays
To determine whether the Bfp60-HIS was enzymatically active, an indirect assay (Jönsson et al., 2004) was used to measure the conversion of plasminogen into plasmin. Bfp60-HIS (10 μg mL−1) was incubated with 4 μg of plasminogen in 200 μL 0.02 M HEPES buffer (pH 7.4) in a 1.5-mL Eppendorf tube. Then, 30 μL of the chromogenic substrate S-2251 (0.45 mM; Val-Leu-Lys-p-nitroalinine dihydrochloride; Chromogenix) was added and incubated at 37 °C. At1-h intervals, a 200-μL sample was transferred to a 96-microwell plate (TPP®) and the colorimetric reaction was measured at 405 nm. Streptokinase (1 U) incubated with plasminogen and plasmin alone were used as positive controls. Plasminogen in HEPES buffer and S-2251 without Bfp60-HIS were used as a negative controls. A similar procedure was also used to test the ability of whole B. fragilis MC2 cells to activate plasminogen by incubating 30 μL of the bacterial cell suspension (3×108 CFU mL−1) with 4 μg of plasminogen before the addition of the chromogenic substrate described above. Staphylococcus aureus ATCC 968 (kindly given by Dr Katia dos Santos Avelar, IMPPG, UFRJ) was used as a positive control for the production of streptokinase, and plasmin alone was used as a positive control. HEPES buffer with an S-2251 substrate was used as a negative control. Data are presented as the means±SEM (SE of the mean). The Student t-unpaired t-test was used, and values with P<0.05 were considered significant.
Results
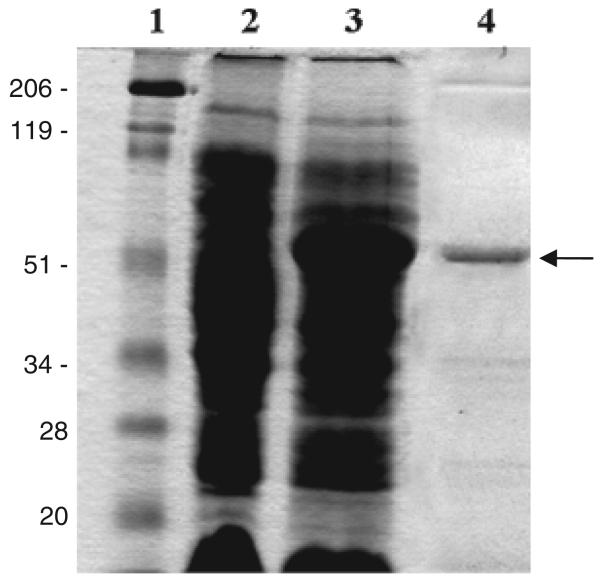
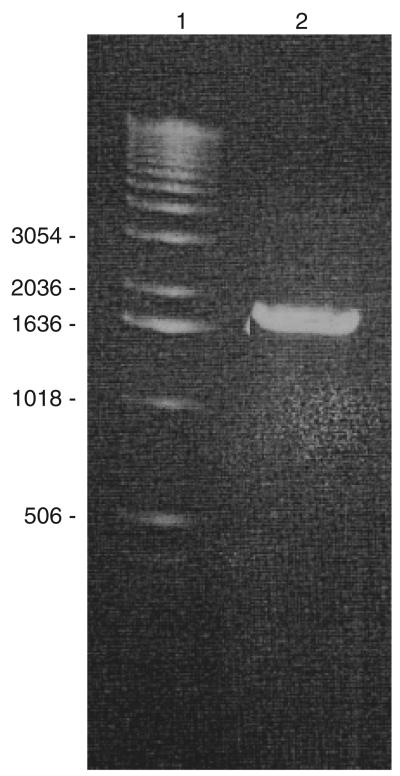
In a previous study, we showed that B. fragilis MC2 was able to strongly adhere to LMN-1 immobilized onto glass cover-slips, but the factors reponsible for this adherence remained unidentified (Ferreira et al., 2006). Thus, in order to characterize the adhesin responsible for LMN-1 recognition, enriched OMP extracts from the B. fragilis MC2 strain were applied to an LMN-1 affinity chromatography column. An OMP protein of c. 60 kDa was eluted at 2 M NaCl (Fig. 1). The isolated protein was identified by peptide massspectroscopy. The amino acid sequence of the peptide fragments matched with 98% similarity to a putative plasminogen-binding 60-kDa protein precursor, named Bfp60, isolated from B. fragilis BE1 strain (Sijbrandi et al., 2005). In order to characterize the protein responsible for adhesion to LMN-1, the bfp60 gene was amplified from B. fragilis MC2 chromosome within a DNA fragment of c. 1700 bp (Fig. 2).
Fig. 1.
SDS-PAGE of the OMPs of the Bacteroides fargilis MC2 strain passed through an affinity column with LMN-1. Lane 1 – molecular weight standards; lane 2 – OMPs of the B. fragilis MC2 strain; lanes 3, 4, 5 and 6 – elutions of the proteins with 0.25, 0.5, 1.0 and 2M NaCl. The arrow (→) indicates the purified fraction.
Fig. 2.
PCR for detection of the gene bfp60 in Bacteroides fragilis MC2 strain. Lane 1 – 1-kb ladder; lane 2 – the amplicon of c.1700bp.
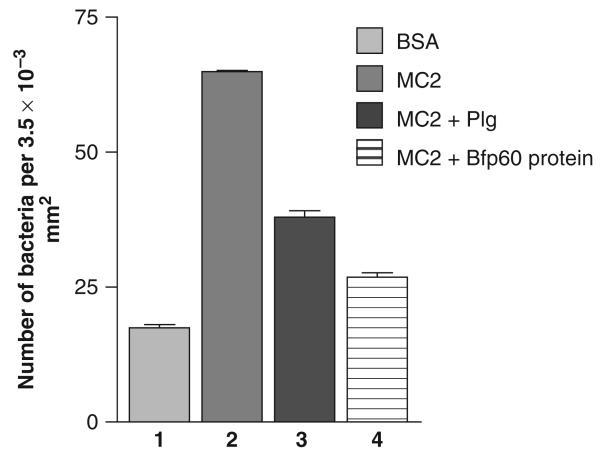
The adhesion of the B. fragilis MC2 strain to immobilized LMN-1 is shown in Fig. 4. When the cells were preincubated with LMN-1 itself or the Bfp60-HP before measuring the adhesion, the binding was significantly lower (P<0.05; P-value: 0.0017). Although plasminogen inhibition seemed to be less strong than Bfp60-HP and LMN-1, this difference was not significant (P>0.05; P–value: 0.128), showing that these proteins, in a certain way, blocked the adhesion of the MC-2 strain to the LMN-1 (Fig. 3, Fig. 4, lanes 3, 4 and 5).
Fig. 4.
Adhesion and inhibition of Bacteroides fragilis MC2 strain to LMN-1. 1 – negative control (2% BSA); 2 – adhesion assay of the strain MC2 to LMN-1; 3, 4 and 5 – inhibition assay of the B. fragilis MC2 strain incubated with LMN-1 (10 μg mL−1), plasminogen (10 μg mL−1) and Bfp60 protein (10 μg mL−1), respectively. All tests were performed in triplicate.
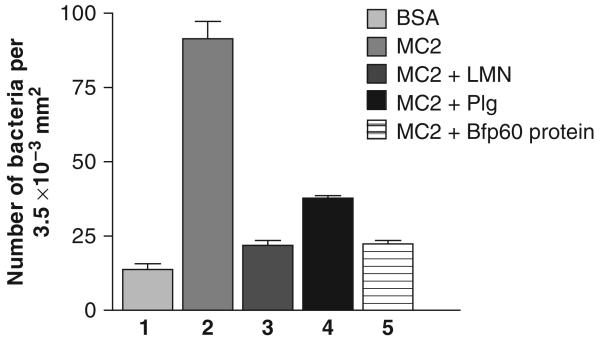
The B. fragilis MC2 strain was also capable of adhering to immobilized plasminogen (Fig. 5), and when the cells were preincubated with plasminogen itself and the Bfp60-HP (Fig. 5, lanes 3 and 4, respectively), both could block the adhesion to plasminogen (P<0.05; P-value: 0.021). Despite the fact that plasminogen did not inhibit the adhesion as strongly as the purified protein, when compared with the Bfp60-HP inhibition, the values were not significant (P>0.05; P-value: 0.11).
Fig. 5.
Adhesion and inhibition assay of Bacteroides fragilis MC2 strain to plasminogen. 1 – negative control (2% BSA); 2 – adhesion assay of the B. fragilis MC2 strain to plasminogen; 3 and 4, inhibition assay of the MC2 strain incubated with 10 μg mL−1 of plasminogen and 10 μg mL−1 of Bfp60-HP, respectively. All tests were performed in triplicate.
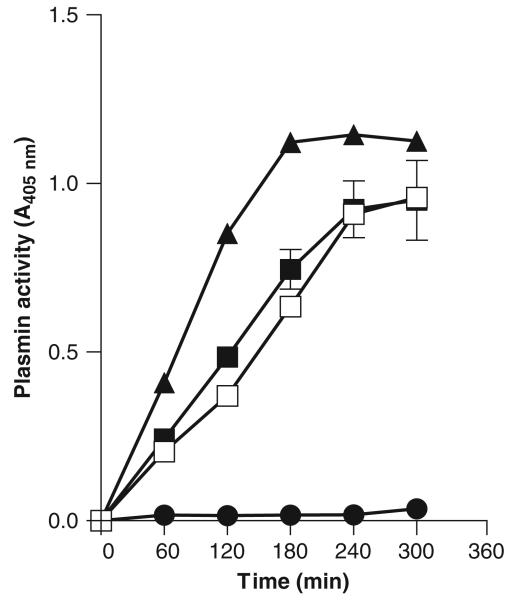
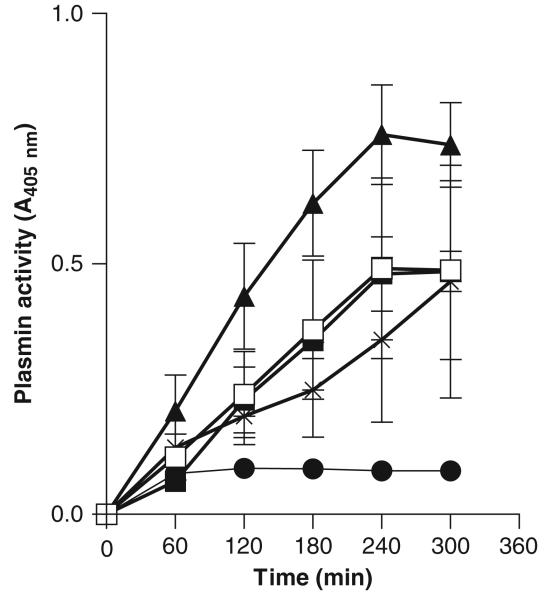
To check whether the Bfp60-HP was capable of converting plasminogen into plasmin, an indirect assay was used. Figure 6 shows the formation of plasmin, measured every hour, with the S-2251 substrate when the protein was incubated with plasminogen. The Bfp60-HP was capable of converting plasminogen into plasmin at a rate similar to the positive controls and was significantly greater than the negative control (P<0.05; P-value: 0.0003). The B. fragilis MC2 strain was also capable of converting the plasminogen into plasmin in whole-cell assays (Fig. 7). Plasmin formation in the B. fragilis MC2 strain was as strong as the positive controls, S. aureus (ATCC 968) strain and streptokinase (P>0.05; P-value: 0.09), but not as good as plasmin itself (P<0.05; P-value: 0.01). Still, the conversion was greater than the negative control (P<0.05; 0.037).
Fig. 6.
Enhancement of plasminogen activation in the presence of Bfp60-HP. Plasmin formation was measured with S-2251 at a Bfp60-HP concentration of 10 μg mL−1. Plasmin with the chromogenic substrate (▲); Bfp60-HP incubated with plasminogen (□); plasminogen in the presence streptokinase (■); negative control plasminogen with HEPES buffer and substrate (●). All tests were performed in duplicate.
Fig. 7.
Plasminogen activation by the Bacteroides fragilis MC2 strain. Plasmin formation by whole cells was measured with S-2251 at a concentration of 108 CFU mL−1. Plasmin with the chromogenic substrate (▲); B. fragilis MC2 strain incubated with plasminogen (□); plasminogen in the presence of streptokinase (■); plasminogen incubated with Staphylococcus aureus ATCC 968 strain (*); negative control plasminogen with HEPES buffer and substrate (●). All tests were performed in duplicate.
Our results showed that the Bfp60 protein effectively reduced the adhesion to LMN-1 as drastically as the LMN-1 itself and that plasminogen also reduced the adhesion by more than 50%. The ability of the B. fragilis MC2 strain to adhere to plasminogen was also tested. The B. fragilis MC2 strain was capable of binding to human plasminogen and again the Bfp60-purified protein inhibited the adhesion better than the plasminogen itself. Taken together, our results show that the bfp60 gene might encode this surface OMP, the Bfp60, which helps to mediate binding to both proteins LMN-1 and plasminogen.
Discussion
Bacteria have several distinct and alternative adhesins for attachment to host cells. Some of these bacterial adhesins may be multifaceted or function in distinct steps to promote tighter binding of bacteria to host cells. When acting collectively, the adhesins define where the pathogen will adhere, colonize and initiate an infection (Dabo et al., 2006).
As a commensal microorganism, B. fragilis must be able to maintain itself in the host intestinal mucosa. On the other hand, as a pathogen, it must be able to attach to the site of the infection, evade the host defense and produce factors that will, most of the time, damage the host tissue. Several virulence factors have been described in B. fragilis (Patrick et al., 1995; Gibson et al., 1998; Krinos et al., 2001; Sears, 2006), but their actual correlation with the pathogenicity of the species is still unclear.
In the mammalian body, tissue barriers are formed especially by the ECM and BM, which have major components, including collagens, laminins, fibronectins, protegly-cans and elastin. The migration through ECM and tissue layers is considered to be a major issue for a bacterial pathogen in its dissemination within the host body (Lähteenmäki et al., 2001), and bacterial proteases are important virulence factors that can improve tissue degradation, bacterial dissemination and/or nutrient release (Travis et al., 1995). Even as a member of the intestinal microbiota, in vitro assays have shown that, when grown under certain conditions (oxidized conditions), B. fragilis can alter its state from commensal to pathogenic, becoming invasive to HeLa cells (Goldner et al., 1993). Adhesion is a complex event and the capacity of this microorganism to express a huge variety of adhesins on its surface (Oyston & Handley, 1990; Patrick & Lutton, 1990) might explain its versatility either as a member of the microbiota or as an opportunistic pathogen. It is also possible that the expression of the adhesins occurs in a strict phase or could be related to an environmental response. Many events in the host may be responsible for the exposition of the basal lamina: (a) it can occur because of the B. fragiles itself (Oyston & Handley, 1990), (b) because of changes in the microbiota of the host and (c) or due to physical traumas (Tally & Ho, 1987). Recently, it has been established that binding and activation of human plasminogen on the surface of bacterial cells may be a common mechanism used by invasive bacteria to facilitate movement through normal tissue barriers (Yarzabal et al., 2000). Indeed, many pathogens interact with host plasminogen, contributing to their pathogenicity (Sun, 2006).
Bacterial OMPs include adhesins that are critical for adherence and colonization (Handerson et al., 1999). Little is known about the possible involvement of surface molecules, especially the OMPs, in the virulence of B. fragilis, but they might play an important role in such crucial events as adhesion, invasion and evasion of host defenses and antimicrobial agents (Patrick & Lutton, 1990; Wexler, 2002). Sijbrandi et al. (2005) isolated an outer membrane protein in B. fragilis capable of recognizing plasminogen, but little has been done in the field to better characterize this interaction. In this article, we performed some assays demonstrating that this adhesin is also involved in the LMN-1 recognition and the conversion of plasminogen into plasmin.
In this study, the protein involved with the LMN-1 adhesion matched to a putative plasminogen binding 60-kDa protein precursor described (Bfp60) in B. fragilis. Hence, it was important to determine whether the Bfp60-HP (recombinant protein) could interfere with the adhesion to LMN-1. The Bfp60 has several predicted tetratricopeptide repeat domains (TRP) that have originally been identified as a protein interaction module in cell division cycle proteins in yeast (Hirano et al., 1990). The TPR motif has since been shown to be ubiquitous, because it is present in a number of proteins that are functionally unrelated and mediates a variety of different protein–protein interactions involved in several biological systems. Depending on the number of tandem TRP motifs and the size of the groove generated, a number of multiple simultaneous interactions with the target proteins could occur (Blatch & Lässle, 1999). This could explain the LMN-1 recognition and plasminogen binding/activation by the Bfp60 protein. Indeed, our results showed that the Bfp60 protein effectively reduced the adhesion to LMN-1 as drastically as the LMN-1 itself and that plasminogen also reduced the adhesion by >50%. The ability of B. fragilis MC2 strain to adhere to plasminogen was also tested. The B. fragilis MC2 strain was capable of binding to human plasminogen and again the Bfp60-purified protein inhibited the adhesion better than the plasminogen itself. Taken together, our results show that the bfp60 gene might encode this surface OMP, the Bfp60, which helps to mediate binding to both proteins LMN-1 and plasminogen.
The number of bacteria, fungi and parasites that are known to recruit and convert plasminogen into plasmin for degradative processes has increased substantially in the last decade. This process usually occurs in two distinct steps. Firstly, the bacteria have to produce receptors that activate the plasminogen complex formation or by proteolysis. Secondly, when the plasminogen is recruited to the bacterial cell surface by these receptors the immobilized plasminogen is converted into plasmin by host-derived activators or the bacteria itself convert it. Either way, cell surface-bound plasmin is exploited by bacteria for proteolytic degradation of components of the ECM (collagens, laminin and fibronectin), BM and host tissues (Travis et al., 1995). This destruction facilitates bacterial translocation and dissemination within the host. The clinical importance of B. fragilis in human anaerobic infections is well known and evidence indicating that the adhesion ability is an important virulence factor has been accumulating (Pruzzo et al., 1989; Patrick et al., 1995, 1999). Several authors have attempted to determine how a bacterium that belongs to the microbiota can eventually become so pathogenic to the host. Since the plasminogen binding and activation on the surface of pathogenic bacteria have been implicated in the invasion of tissues, we decided to test whether the Bfp60 itself and the B. fragilis MC2 strain were capable of converting plasminogen into plasmin. Our results show that the purified protein is capable of converting the plasminogen into plasmin as good as 1 U of streptokinase. When the B. fragilis MC2 strain was tested in the whole-cell assay, the bacterium was capable of forming as much plasmin as the S. aureus ATCC 968. We believe that the Bfp60-purified protein formed more plasmin than the B. fragilis MC2 strain, because the concentration of the bacteria used in the test (3×108 CFU mL−1) did not reach the same concentration as the protein used in the assay (10 μg mL−1), even though our results proved that both the purified protein and the strain were capable of converting plasminogen into plasmin.
Plasminogen is an abundant circulating precursor that can be activated by several pathogens. The resulting plasmin transforms the bacteria into a proteolytic organism using the host-derived system. The bacterial infection can also influence the production of the tPA and plasmin inhibitors (Lähteenmäki et al., 2005). Hence, the activation of plasminogen by B. fragilis can help this bacteria to cross through several barriers, such as the intestinal epithelial and the ECM, and reach the blood stream. We believe that the presence of this protein in the species can enhance the virulence of this organism. In this regard, we analyzed 51 B. fragilis strains isolated from different clinical infections and we found the presence of the bfp60 gene in all of them (data not shown).
In conclusion, we presented in vitro evidence for a pathogenic function of the plasminogen receptor in promoting adherence to laminin and also the conversion of plasminogen into plasmin by B. fragilis. Adherence to laminin and plasminogen activation are extremely important in the metastasis of tumor cells and the same behavior is exhibited by a number of pathogens. Our results encourage us to continue our studies in this field, and further studies are being performed for further characterization of this adhesin to highlight the pathogenicity of B. fragilis.
Acknowledgements
The authors would like to thank Joaquim Santos Filho and Anita Parker for their technical assistance. This work has been supported by the following Brazilian agencies: CAPES, CNPq, PRONEX and FAPERJ and the NIH/NIAID (AI068659).
References
- Blatch GL, Lässle M. The tetratricopeptide repeat: a structutral motif mediating protein–protein interaction. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bölin I, Norlander L, Wolf-Watz H. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with virulence plasmid. Infect Immun. 1982;17:506–512. doi: 10.1128/iai.37.2.506-512.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MD, Lottenberg R. Plasminogen activation by invasion human pathogens. Thromb Haemostasis. 1997;77:1–10. [PubMed] [Google Scholar]
- Brook I, Frasier EH. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J Med Microbiol. 2000;49:827–830. doi: 10.1099/0022-1317-49-9-827. [DOI] [PubMed] [Google Scholar]
- Dabo SM, Confer AW, Saliki JT, Anderson BE. Binding of Bordetella henselae to extracelullar molecules: identification of potential adhesins. Microb Pathogenesis. 2006;41:10–20. doi: 10.1016/j.micpath.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ferreira EO, Lobo LA, Petrópolis DB, Avelar KES, Ferreira MC, Silva Filho FC, Domingues RMCP. A Bacteroides fragilis surface glycoprotein mediates the interaction between the bacterium and the extracellular matrix component laminin-1. Res Microbiol. 2006;157:960–966. doi: 10.1016/j.resmic.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Finegold SM. Overview of clinically important anaerobes. Clin Infect Dis. 1995;20:205–207. doi: 10.1093/clinids/20.supplement_2.s205. [DOI] [PubMed] [Google Scholar]
- Gibson FC, Onderdonk AB, Kasper DL, Tzianabos AO. Cellular mechanisms of intraabdominal abscess formation by Bacteroides fragilis. J Immunol. 1998;160:5000–5006. [PubMed] [Google Scholar]
- Goldner M, Coquis-Rondon M, Carlier JP. Effect of growth of Bacteroides fragile at different redox levels on potential pathogenicity in a HeLa cells system: demonstration by confocal laser scanning microscopy. Zentralbl Bakteriol. 1993;278:529–540. doi: 10.1016/s0934-8840(11)80824-8. [DOI] [PubMed] [Google Scholar]
- Handerson B, Wilson M, McNab R, Lax AJ. Cellular Microbiology: Bacteria–Host Interactions in Health and Disease. Wiley; New York: 1999. [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essencial repeats in Schizosaccharomyces pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- Jönsson K, Guo BP, Monstein HJ, Mekalanos JJ, Kronvall G. Molecular cloning and characterization of two Helicobacter pylori genes coding for plasminogen-binding proteins. PNAS. 2004;101:1852–1857. doi: 10.1073/pnas.0307329101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousiemies-Somier H, Summanen P, Citron DM, Baron EJ, Wexler HM, Finegold SM. Wadsworth Anaerobes Bacteriology Manual. 6th edn. Star Publishing Company; Belmont, CA: 2002. pp. 39–63. [Google Scholar]
- Kern PA, Schotz MC. An enzyme linked immunoassay for lipoprotein lipase. Anal Biochem. 1987;166:25–35. doi: 10.1016/0003-2697(87)90541-0. [DOI] [PubMed] [Google Scholar]
- Krinos CM, Coyne MJ, Weinacht XG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- Laemmili UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lähteenmäki K, Virkola R, Riitta P, Kuusela P, Kukkonen M, Korhonen TK. Bacterial Plasminogen receptors: in vitro evidence for a role in degradation of the mammalian extracelullar matrix. Infect and Immun. 1995;63:3659–3664. doi: 10.1128/iai.63.9.3659-3664.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lähteenmäki K, Kukkonen M, Korhonen TK. The Pla protease/adhesin of Yersinia pestis mediates bacterial invasion into human endothelial cells. FEBS Lett. 2001;504:69–72. doi: 10.1016/s0014-5793(01)02775-2. [DOI] [PubMed] [Google Scholar]
- Lahteenmaki K, Kuusela P, Korhonen TK. Bacterial plasminogen activators and receptors. FEMS Microbiol Rev. 2001;25:531–552. doi: 10.1111/j.1574-6976.2001.tb00590.x. [DOI] [PubMed] [Google Scholar]
- Lähteenmäki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 2005;13:79–85. doi: 10.1016/j.tim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Lissitsky JC, Cantou P, Martin PM. Heterotrimeric configuration is essential to the adhesive function of laminin. J Cell Biochem. 2004;48:141–149. doi: 10.1002/jcb.240480206. [DOI] [PubMed] [Google Scholar]
- Mökänen T, Tyynelä J, Helin J, Kalkkinen N, Kuusela P. Enhanced activation of bound plasminogen on Staphylococcus aureus by staphylokinase. FEBS Lett. 2002;517:72–78. doi: 10.1016/s0014-5793(02)02580-2. [DOI] [PubMed] [Google Scholar]
- Oyston PC, Handley PS. Surface structures, haemagglutination and cell surface hydrophobicity of Bacteroides fragiles strains. J Gen Microbiol. 1990;136:941–948. doi: 10.1099/00221287-136-5-941. [DOI] [PubMed] [Google Scholar]
- Patrick S, Lutton DA. Outer membrane proteins of Bacteroides fragilis grown in vivo. FEMS Microbiol Lett. 1990;71:1–4. doi: 10.1016/0378-1097(90)90022-i. [DOI] [PubMed] [Google Scholar]
- Patrick S, Lutton DA, Crockard AD. Immune reactions of Bacteroides fragilis populations with three different types of capsule in a model of infection. Microbiology. 1995;141:1969–1976. doi: 10.1099/13500872-141-8-1969. [DOI] [PubMed] [Google Scholar]
- Patrick S, McKenna JP, O'Hagan S, Dermotte E. Comparasion of the haemaggluting and enzymic activites of Bacteroides fragilis whole cells and outer membrane vesicles. Microb Pathogenesis. 1996;20:191–202. doi: 10.1006/mpat.1996.0018. [DOI] [PubMed] [Google Scholar]
- Patrick S, Gilpin D, Stevenson L. Detection of intra-strain antigenic variation of Bacteroides fragilis surface polysaccharides by monoclonal antibody labeling. Infect Immun. 1999;67:4346–4351. doi: 10.1128/iai.67.9.4346-4351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzzo C, Guzman CA, Dainelle B. Incidence of hemagglutination activity among pathogenic and nonpathogenic Bacteroides fragilis. FEMS Microbiol Lett. 1989;59:113–118. doi: 10.1016/0378-1097(89)90469-2. [DOI] [PubMed] [Google Scholar]
- Pumbwe L, Skilbeck CA, Wexler HM. The Bacteroides fragilis cell envelope: quarterback, linebacker, coach – or all three? Anaerobe. 2006;12:211–220. doi: 10.1016/j.anaerobe.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sears CL. The toxins of Bacteroides fragilis. Toxicon. 2006;39:1737–1746. doi: 10.1016/s0041-0101(01)00160-x. [DOI] [PubMed] [Google Scholar]
- Shah N. The procaryotes. In: Balows A, Truper HG, Dworkin M, Harder W, Scheleifer KH, editors. The Genus Bacteroides Fragilis and Related Species. 2nd edn Springer-Verlag; New York: 1999. pp. 3593–3607. [Google Scholar]
- Sijbrandi R, Den Blaauwen T, Tame JR, Oudega B, Luirink J, Otto BR. Characterization of an iron-regulated alpha-enolase of Bacteroides fragilis. Microbes Infect. 2005;7:9–18. doi: 10.1016/j.micinf.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Sun H. The interaction between pathogens and the host coagulation system. Physiology. 2006;21:281–288. doi: 10.1152/physiol.00059.2005. [DOI] [PubMed] [Google Scholar]
- Tally FP, Ho JL. Management of patients with intraabdominal infection due to colonic perforation. Curr Clin Top Infect Dis. 1987;8:266–295. [PubMed] [Google Scholar]
- Travis J, Potempa J, Maeda H. Are bacterial proteinases pathogenic factors? Trends Microbiol. 1995;3:405–407. doi: 10.1016/s0966-842x(00)88988-x. [DOI] [PubMed] [Google Scholar]
- Ullberg M, Kuusela P, Kristiansen BE, Kronvall G. Binding of plasminogen to Neisseria meningitidis and Neisseria gonorrhoeae and formation of surface-associated plasminin. J Infect Dis. 1992;166:1329–1334. doi: 10.1093/infdis/166.6.1329. [DOI] [PubMed] [Google Scholar]
- Wexler HM. Outer membrane pore-forming proteins in Gram negative anaerobic bacteria. Clin Infect Dis. 2002;35:S65–S71. doi: 10.1086/341923. [DOI] [PubMed] [Google Scholar]
- Wu S, Kuei-Chung L, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. P Natl Acad Sci USA. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarzabal A, Avilán L, Hoelzl K, de Munoõz M, Puig J, Kansau I. A study of the interaction between Helicobacter pylori and components of the human fibrinolytic system. Braz J Med Biol Res. 2000;33:1015–1021. doi: 10.1590/s0100-879x2000000900004. [DOI] [PubMed] [Google Scholar]