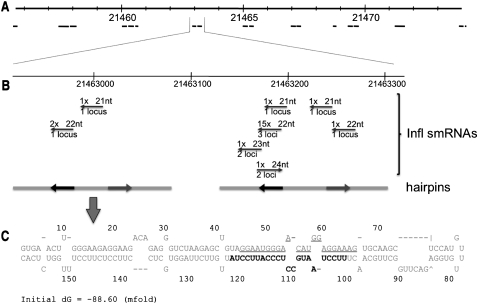
Figure 5.
The genomic origins of the motif-targeting small RNAs, miR2118, and potential folding of transcripts containing these miRNAs. (A) The ∼20-kb region that codes for most of the motif-targeting small RNAs. This region contains nine degenerate repeats each comprised of two to three inverted repeats. Inverted repeats are indicated by black lines below. (B) An enlargement of the indicated region. The small RNAs potentially derived from this region are displayed. The polarity of the small RNAs is indicated by the arrows, and the number of times the small RNA was sequenced and the size of the small RNA are indicated above the arrow. The number of loci potentially encoding each small RNA is shown below the arrow. The positions of the motif-targeting small RNA (black) and the motif-targeting small RNA* (gray) are shown on the gray line representing the inverted repeat. The left inverted repeat contains the motif-targeting 22-nt small RNA (smRNA165692) while the right inverted repeat contains a 22-nt motif-targeting small RNA (smRNA165598) that maps to an additional two locations in the overall region. (C) The predicted hairpin structure of the indicated inverted repeat. The motif-targeting small RNA, miR2118, is shown in black and the motif-targeting RNA*, miR2118*, is underlined.