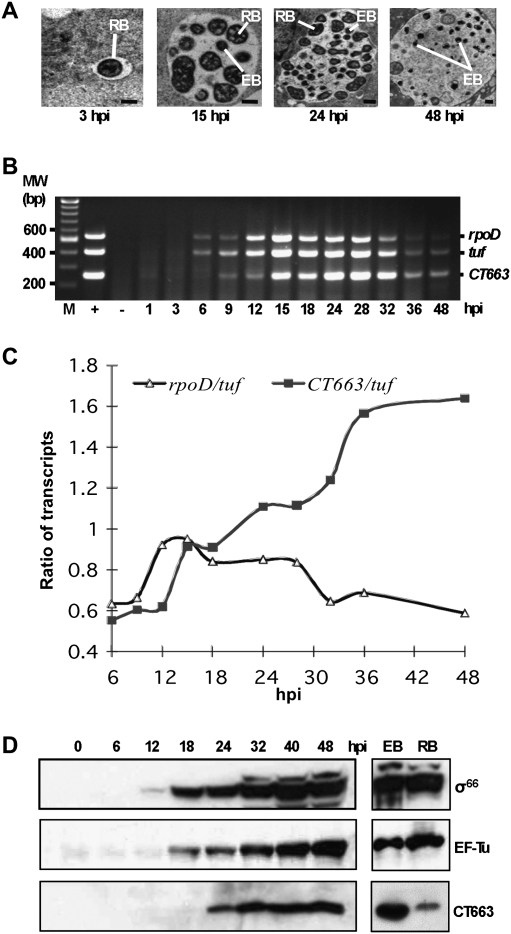
Figure 5.
C. trachomatis developmental gene expression. (A) Transmission electron micrographs of C. trachomatis serovar F within an inclusion. C. trachomatis-infected cells were sampled at different time points post-infection as indicated. Representatives of the two major C. trachomatis forms within the inclusion, the EB and the RB, are labeled (bar, 0.5 μm). (B) Results of multiplex RT–PCR analysis. Ethidium bromide-stained agarose gel showing the developmental expression pattern of the CT663, tuf, and rpoD transcripts harvested from C. trachomatis-infected L929 cells. Genomic DNA extracted from purified C. trachomatis EBs was used as a positive control to show that the three primer pairs amplified their targets with similar efficiencies (lane +) when used in this multiplex format. RNA isolated from L929 cells uninfected with C. trachomatis was used as negative control (lane −). A fixed quantity of total RNA (host cell RNA + C. trachomatis RNA) isolated at the indicated times in hours post-infection (hpi) was treated with DNase I and was subjected to RT–PCR. The products of the RT–PCRs migrated at the expected sizes. (C) Graph showing the ratio of rpoD and CT663 transcripts to the tuf transcript at different time points during the C. trachomatis developmental cycle as revealed by the RT–PCR assay in B. Note that there is a general decrease in transcript levels that occurs late in the developmental cycle due to the conversion of the metabolically active RB form to the relatively inactive EB form; nevertheless, when compared with the tuf transcript there is relatively more CT663 transcript and relatively less rpoD transcript than in the earlier stages of infection. (D) Developmental protein synthesis pattern for CT663, EF-Tu (tuf gene product), and σ66 (rpoD gene product) harvested from C. trachomatis-infected L929 cells. A normalized amount of total protein (host cell protein + C. trachomatis protein) isolated from C. trachomatis-infected L929 cells at the indicated time points in hours post-infection (hpi) was separated by 12% SDS-PAGE and probed with antibodies specific to CT663, σ66, or EF-Tu. A normalized amount of protein isolated from purified EBs (lane EB) and purified RBs (lane RB) was also immunoblotted as indicated. All protein bands migrated at the expected sizes.