Table 1. Crystal parameters, data collection and structure refinement.
Values in parentheses are for the highest resolution bin.
| Data processing | |
| Space group | H3 |
| Unit-cell parameters (, ) | a = b = 140.610, c = 46.050, = = 90, = 120 |
| Resolution range () | 43.072.38 (2.472.38) |
| Unique reflections | 12995 (1089) |
| Unique reflections after truncation | 11503 (406) |
| Completeness (%) | 94.7 (80.0) |
| Completeness after truncation (%) | 83.2 (51.4) |
| I/(I) | 21.72 (5.40) |
| R merge † (%) | 6.1 (20.9) |
| Redundancy | 3.7 |
| Refinement statistics | |
| Resolution range () | 20.002.38 (2.472.38) |
| R factor‡/R free § (%) | 22.3/26.5 (33.5/35.7) |
| No. of protein atoms | 1896 |
| No. of water atoms | 114 |
| R.m.s.d.¶ bond length () | 0.011 |
| R.m.s.d. bond angles () | 1.245 |
| Average of B factors (2) | 34.54 |
| Ramachandran plot†† | |
| Most favoured (%) | 98.1 |
| Additional allowed (%) | 1.9 |
| Outliers (%) | 0 |
| PDB code | 3hia |
R
merge = 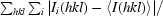
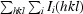 , where I
i(hkl) is the intensity of the ith observation of reflection hkl and I(hkl) is the mean value for reflection hkl; summations are over all reflections.
, where I
i(hkl) is the intensity of the ith observation of reflection hkl and I(hkl) is the mean value for reflection hkl; summations are over all reflections.
R factor = 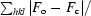
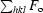 , where F
o and F
c are the observed and calculated structure-factor amplitudes, respectively.
, where F
o and F
c are the observed and calculated structure-factor amplitudes, respectively.
R free was calculated with 5% of the data excluded from the refinement.
Root-mean-square deviation from ideal values.
Categories were defined by MolProbity.
