Table 1. Crystal and refinement parameters for the Z isomer of 2,4-diamino-5-[2-(2′-methoxyphenyl)propenyl]-furo[2,3-d]pyrimidine as a ternary complex with human DHFR and NADPH.
Values in parentheses are for the highest resolution shell.
| PDB code | 3gyf |
| Space group | P212121 |
| Unit-cell parameters (Å) | |
| a | 39.91 |
| b | 57.48 |
| c | 74.97 |
| Beamline | SSRL 9-2 |
| Resolution (Å) | 45.64–1.70 (1.79–1.70) |
| Wavelength (Å) | 0.979 |
| Rmerge† (%) | 0.06 (0.22) |
| Completeness (%) | 97.2 (93.9) |
| Observed reflections | 196156 (25778) |
| Unique reflections | 19108 (2656) |
| I/σ(I) | 22.2 (9.0) |
| Multiplicity | 10.3 (9.7) |
| Refinement and model quality | |
| Resolution range (Å) | 31.40–1.70 (1.70–1.79) |
| No. of reflections | 18089 (1310) |
| R factor‡ | 21.3 (26.0) |
| Rfree factor§ | 26.4 (29.3) |
| Total protein atoms | 1626 |
| Total water atoms | 126 |
| Average B factor (Å2) | 24.4 |
| R.m.s. deviation from ideal | |
| Bond lengths (Å) | 0.023 |
| Bond angles (°) | 2.03 |
| E.s.d. from Luzzati plot (Å) | 0.22 |
| Ramachandran plot | |
| Most favored regions (%) | 95.7 |
| Additional allowed regions (%) | 2.7 |
| Generously allowed regions (%) | 1.6 |
| Disallowed regions (%) | 0.0 |
R
merge = 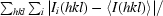
 , where 〈I(hkl)〉 is the mean intensity of a set of equivalent reflections.
, where 〈I(hkl)〉 is the mean intensity of a set of equivalent reflections.
R factor = 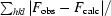
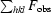 , where F
obs and F
calc are observed and calculated structure-factor amplitudes.
, where F
obs and F
calc are observed and calculated structure-factor amplitudes.
The R free factor was calculated as for the R factor but for a random 5% subset of all reflections.
