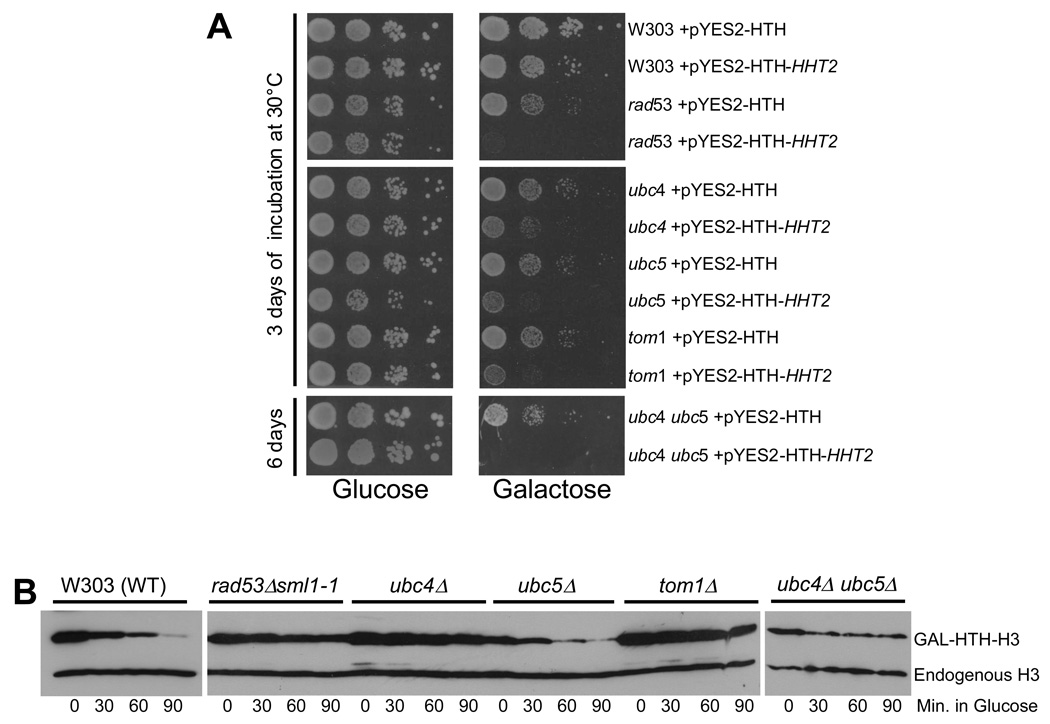
Figure 4. Identification of the putative E2 and E3 enzymes involved in the degradation related ubiquitylation of histones.
(A) Yeast strains lacking Ubc4, Ubc5 and Tom1 are sensitive to histone overexpression. Wild type W303 and congenic strains carrying deletions of genes encoding E2 or E3 enzymes were transformed with either a plasmid encoding galactose inducible, HIS10-TEV-HA (HTH) tagged histone H3 (pYES2-HTH-HHT2) or the empty vector (pYES2-HTH)9. The resulting transformants were grown overnight in minimal medium lacking uracil (to select for the plasmids) and containing 2% raffinose as carbon source. 10-fold serial dilutions of each strain were plated on minimal media minus uracil, with either glucose or galactose as carbon source. The plates were incubated for 3 days at 30°C, except for the slow growing ubc4 ubc5 double mutants which were incubated for 6 days. The rad53 deletion strain carries the sml1-1 null allele to suppress the lethality due to the loss of the essential function of Rad53.
(B) Disruption of the UBC4, UBC5 and TOM1 genes causes defects in the degradation of overexpressed histones. In vivo histone degradation assay with wild type (W303) and congenic mutant strains carrying the pYES2-HTH-HHT2 plasmid were performed as described in Methods. Cells were harvested every 30 minutes after switching to glucose. The levels of HTH-tagged H3 and chromosomal histone H3 were quantitated by Western blotting using the H3-C antibody9.