Figure 6.
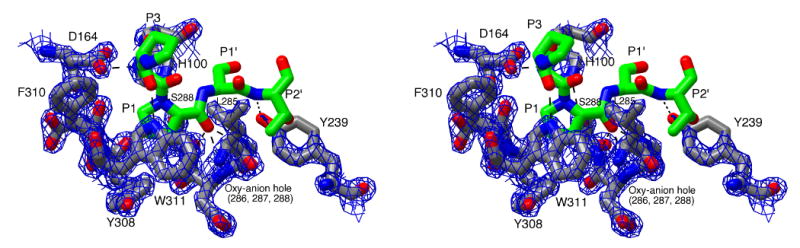
Stereoview of the active site of IgAP illustrating the recognition of the hinge peptide. The P3-P2′ region (229P-S-P-S-T233) of the hinge peptide was manually modeled into the active site of IgAP based upon the position of the equivalent region in the elastase-turkey ovomucoid inhibitor complex and is colored by atom type with the carbon atoms being rendered in green. The catalytic triad and residues of IgAP involved in substrate recognition are colored by atom type with the carbon atoms being rendered in grey. Potential hydrogen bonds between the substrate peptide and the enzyme and members of the catalytic triad (D164, H100, S288) are illustrated by dashed lines. 2FO-FC density for the protein rendered at 2.5σ is shown as a blue mesh.
