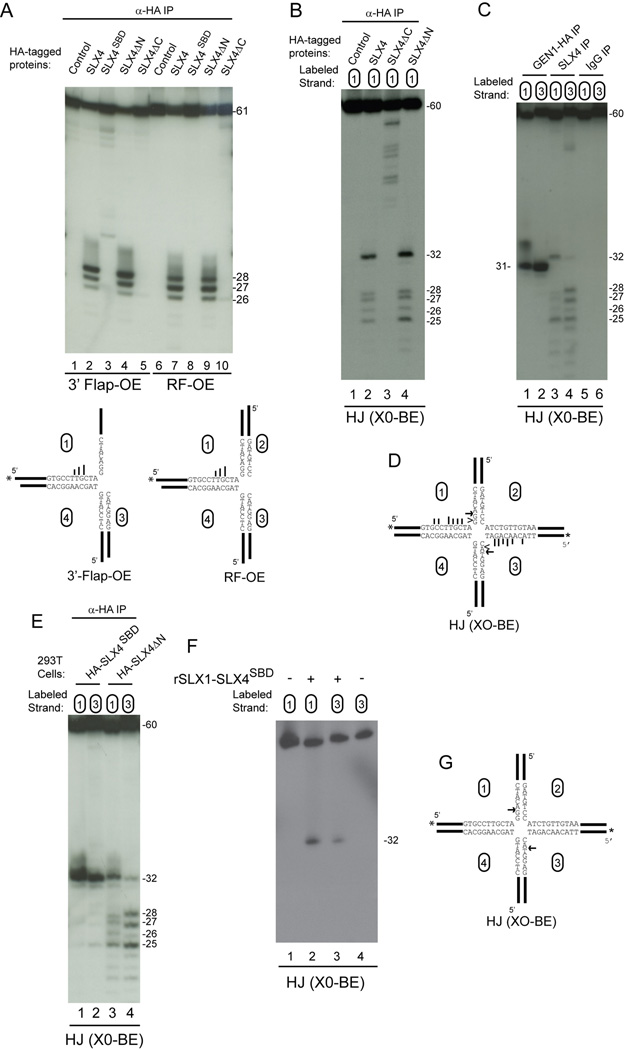
Figure 6. Cleavage specificity of SLX4 and SLX1-SLX4SBD complexes.
(A) The indicated SLX4 complexes purified from 293T cells were incubated with radiolabeled substrates (30 min) prior to 12% polyacrylamide-urea gel electrophoresis and autoradiography. In the structural model (bottom) longer bars represent more efficient cleavage.
(B) HJ cleavage activity of SLX4 maps to the C-terminus containing the MUS81-EME1 and SLX1 binding sites. The indicated SLX4 immune complexes derived from 1.5 mg of 293T cell extract were incubated with HJ (X0-BE) labeled on strand 1 (30 min) and the products separated on denaturing gels.
(C) Symmetrical cleavage of HJ (X0-BE) by SLX4 complexes and GEN1-HA. The indicated immune complexes were incubated with HJ (X0-BE) radiolabeled on either strand 1 or strand 3 and products resolved on a denaturing gel prior to autoradiography.
(D) Major sites of cleavage observed with SLX4 (arrow and bars) or GEN1 (arrowhead) complexes.
(E–G) Symmetrical cleavage of HJ (X0-BE) by SLX1-SLX4SBD complexes. The indicated SLX4 complexes purified from 293T cells (panel E) or bacteria (panel F) were incubated with HJ (X0-BE) radiolabeled on strand 1 or strand 3 and products resolved on a denaturing gel prior to autoradiography. In panel G, the major site of HJ (X0-BE) cleavage by SLX1-SLX4SBD is indicated by the arrow.