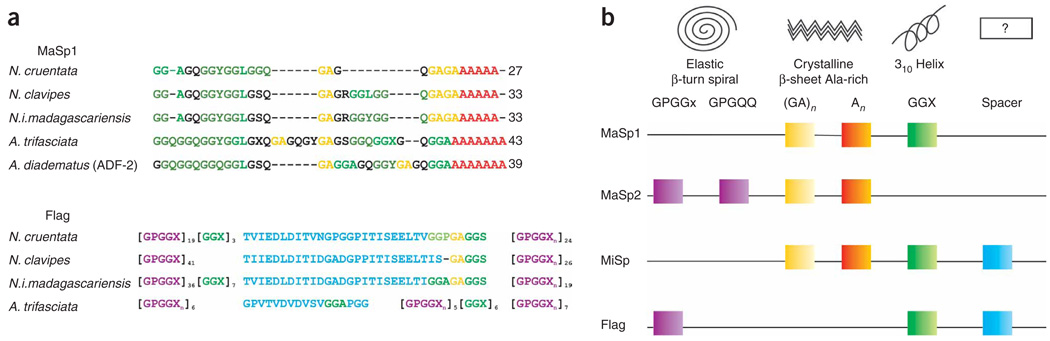
Figure 1.
The molecular structure of orb-weaver spiders’ silk proteins. Alignment of the consensus repetitive sequences of (a) major ampullate (MaSp 1) and flagelliform (Flag) silk proteins (adapted from ref. 20). Structural amino-acid motifs found consensus repeats of spider silk proteins (b). The square-colored boxes indicate that the structural motif is part of the silk protein. To determine which silk proteins contain which modules, read the line that connects the boxes from left to right, starting with the silk name. The empty box marked ’?’ indicates that the secondary structures of these ’spacer’ motifs are unknown. (Adapted from ref. 21.) MaSp1 or MaSp2: major ampullate spidroin 1 or 2; MiSp: minor ampullate spidroin; Flag: flagelliform protein.