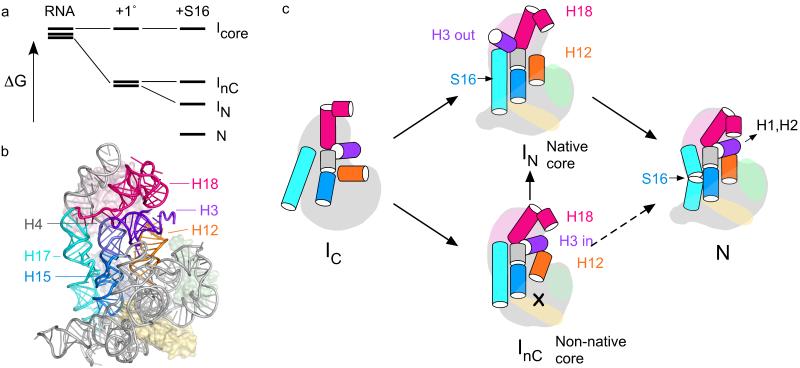
Figure 7. Model for assembly of the 30S 5′ domain.
(a) Free energy diagram illustrating how selective stabilization of a native-like intermediate by S16 depopulates competing I’s and results in more cooperative assembly, 1°, S4+S17+S20. (b) Helices that switch conformation when bound by S16 (blue surface) are highlighted in a structure of the 30S ribosome (2avy). (c) An ensemble of partly folded RNA (IC) containing different configurations of core helices are bound and further stabilized by primary assembly proteins S4, S17 and S20 (pale pink, green and yellow). In the absence of S16, a native-like (IN) and a non-native intermediate (InC) have similar free energies and are both formed. S16 (light blue) preferentially stabilizes IN, resulting in depopulation of InC and a smoother transition to the native RNP (N). Long-distance communication between the binding site of S16 in H17 (cyan) and H15 (blue) and helix 3 (purple) stabilizes the helix 18 pseudoknot (pink) in the 30S decoding center. Equilibrium data do not distinguish paths from InC to N (dashed arrows).