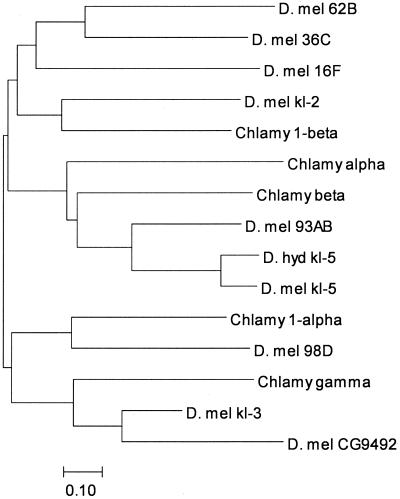
Figure 4.
Dynein neighbor-joining tree. Inferred amino acid sequences were aligned with clustalw (33), and the MEGA2 package (34) was used to construct this neighbor-joining tree. The clustering of kl-3 with Chlamydomonas γ-dynein and the autosomal paralog CG9492 is clear, as is the clustering of kl-5 with Chlamydomonas β-dynein. The autosomal paralog Dhc 93AB. kl-2 clusters with the Chlamydomonas inner arm dynein 1β. Because of its short length, CG9068 (the autosomal paralog of kl-2) could not be included in the phylogeny, but both proteins are closely related, having an amino acid identity of 53%. Accession numbers: kl-5, AAF21041; D. hydei kl-5, AAC35745; Dhc93AB, AAF55834; Dhc16F, AAF48792; Dhc62B, AAF47564; Dhc98D, AAF56793; CG9492, AAF54422; CG9068, AAF58022; Dhc36C, AAF53626; Chlamydomonas β, T08030; Chlamydomonas γ, T08044; Chlamydomonas α, AAA57316; Chlamydomonas 1a, CAB56598; Chlamydomonas 1β, CAB99316. The bar indicates the number of amino acids substitutions per site. D. mel, D. melanogaster; Chlamy, Chlamydomonas; D. hyd, D. hydei.