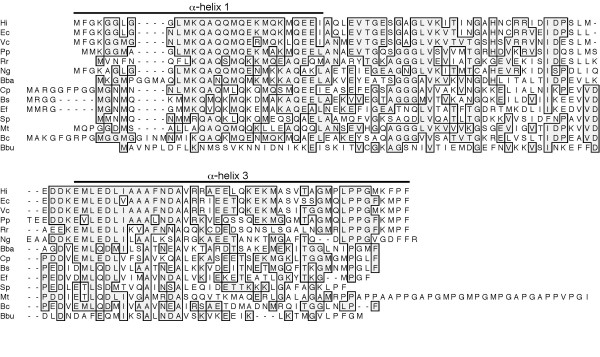
Figure 1.
Alignment of the predicted amino acid sequences of YbaB/EbfC orthologs of H. influenzae (Hi), E. coli (Ec), Vibrio cholerae (Vc), Pseudomonas putida (Pp), Rickettsia rickettsiae (Rr), Neisseria gonorrhoeae (Ng), Bdellovibrio bacteriovorus (Bba), Clostridium perfringens (Cp), Bacillus subtilis (Bs), Enterococcus faecalis (Ef), Streptococcus pneumoniae (Sp), Mycobacterium tuberculosis (Mt), Bacteroides capillosus (Bc), and B. burgdorferi (Bbu). Identical amino acids are boxed and shaded. Amino acid residues of YbaBEc and YbaBHi that comprise αlpha-helices 1 and 3 of their determined protein structures are identified.