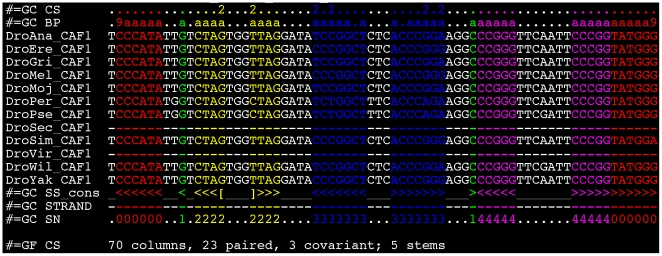
Figure 3. Recovery of a tRNA (FlyBase gene identifier FBgn:0050220) on chromosome 2R.
We recover the four stems of the classic cloverleaf structure, as well as a spurious single base-pair annotated as stem 1 (green). The 5′ boundary is exactly recovered and the 3′ boundary is 2 nt shorter than the FlyBase annotation. Note that stems 2 and 3 (yellow and blue) have, respectively, one and two compensatory mutations. If a base pair exhibits compensatory mutations, the “CS” row shows the count of distinct canonical base-pairs in the columns. The “BP” column shows how many sequences contain a canonical base pair in the consensus structure (“a” = 10). The “SS_cons” row indicates the ML consensus secondary structure predicted by our model; colors of nucleotides and numbers in the “SN” row indicate the stems of this predicted structure. Figure produced with colorstock, described by [51]. The alignment is in the Stockholm file format used by RFAM [37].