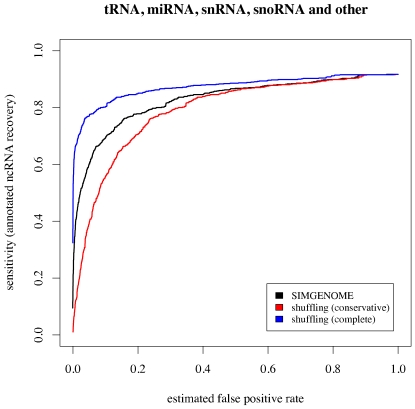
Figure 8. Receiver Operator Characteristic (ROC) curves for the ClosingBp grammar (see “Design of ncRNA gene model”), using our simulated data and two modes of shuffling to generate true-negative datasets.
True-positive datasets were taken from the PECAN alignments of twelve Drosophila genomes based on all annotated non-ribosomal ncRNAs in FlyBase Release 5.4 of the D. melanogaster genome. False-positive estimates from a shuffling-based approach depended strongly on the amount of shuffling. We ran the shuffle-aln.pl script provided with the Vienna RNA package [52] in “conservative” and “complete” modes to create shuffled alignments of all annotated ncRNAs in D. melanogaster.