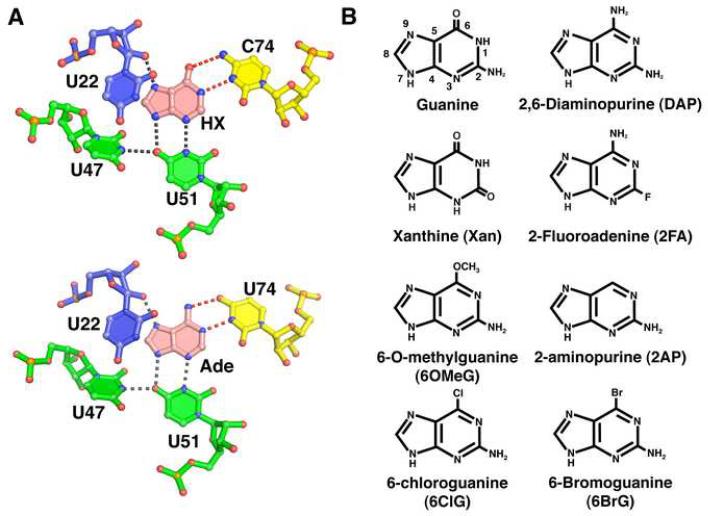
Figure 1. Ligand binding site of the purine riboswitch and chemicals.
(A) Details of hypoxanthine (top, PDB 1U8D) and adenine (bottom, PDB 1Y26) bound to the guanine and adenine riboswitches, respectively. The specificity pyrimidine (C74 or U74, yellow) forms hydrogen bonds (red dashed lines) to the Watson-Crick face of the ligand (pink), while U51, U47, and U22 interact with the other faces of the purine nucleobase. Residues are colored according to their strand position in the junction region. Note that the hydrogen bonding pattern for ligand recognition is identical. (B) Chemical structures of purine derivatives characterized in this study.