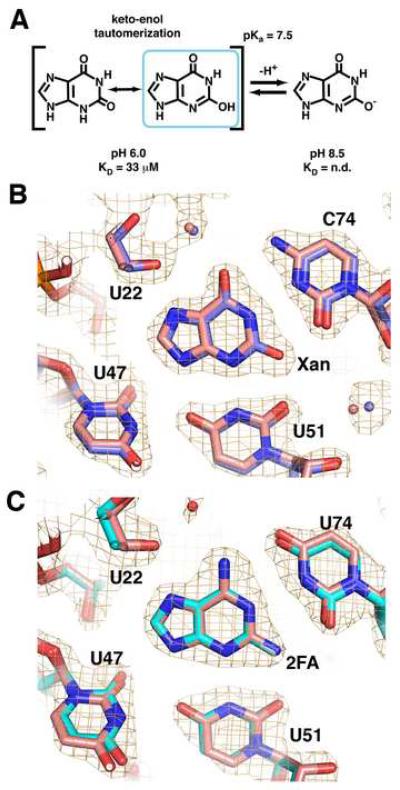
Figure 2. 2-position purine derivatives bound to the purine riboswitch.
(A) Chemical structure of xanthine, emphasizing the keto-enol tautomerization of the N3/O2 atoms and the deprotonation of the enolic proton. (B) Simulated annealing omit map of the X:GR complex in which xanthine, U51, and C74 are removed and contoured at 1.0σ orange cage); the X:GR model is depicted with blue carbon atoms. Superposition of the xanthine-bound (blue) and hypoxanthine-bound (pink) guanine riboswitches shows that base positioning within the binding pocket is unaltered. (C) Simulated-annealing omit map of the 2FA:GRA complex with 2FA, U51, and C74 omitted and contoured at 1.0σ model of 2FA:GRA complex is pink). Superposition of the 2FA-bound (cyan) and 2,6-diaminpurine-bound (pink) GRA RNAs reveals the binding pocket is also unaltered.