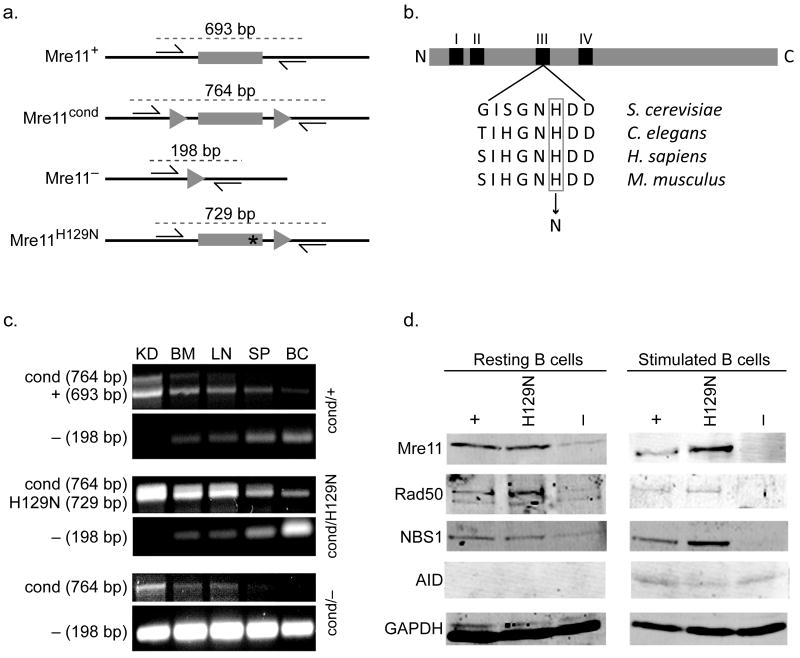
Figure 1. Mre11 deficiencies in the B lymphocyte lineage.
(a) Four germline mouse alleles of Mre11: Mre11 wild type (Mre11+), Mre11 conditional (Mre11cond), Mre11 null (Mre11−), and Mre11 deficient in nuclease activities (Mre11H129N). Mre11 exon 5 (gray rectangle), intronic sequence (black line), LoxP sites (triangles), histidine to asparagine mutation at amino acid 129 (asterisk). Base pair (bp) numbers indicate allele specific PCR products resulting from the two primers depicted (arrows)9. (b) Location of the invariant active site histidine within nuclease motif III. Mouse histidine 129 was changed to asparagine as described9. (c) PCR analyses distinguishing the four Mre11 germline alleles. Conversion of Mre11cond to Mre11− is detected only in sites containing substantial numbers of B lymphocytes (BM - bone marrow, LN - lymph node, SP - whole spleen, BC - enriched B cell population from spleen), but not in kidney (KD). Primers used depicted in (a). (d) Western blot analyses of MRN components and AID in splenic B lymphocytes from mice harboring one allele of CD19-Cre. GAPDH was used as a protein loading control. Left - resting B cells, right - B cells stimulated to undergo class switch recombination for 4 days in culture. Each mouse contains the Mre11 allele indicated, and a second allele which is Mre11− in the B lineage (Mre11cond elsewhere).