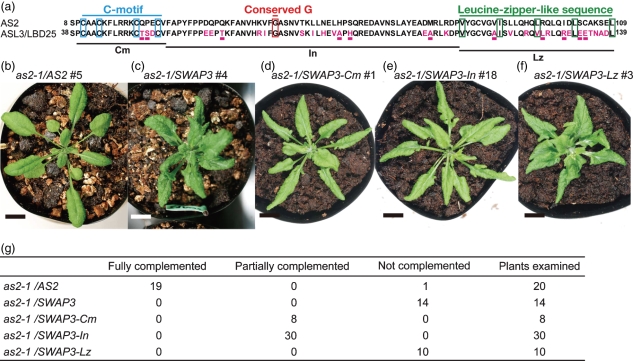
Figure 6.
Replacement of three regions in the AS2/LOB domain of AS2 with the corresponding regions of ASL3/LBD25. (a) Comparison of the amino acid sequences of the AS2/LOB domains of AS2 and ASL3/LBD25. The sequence from residue 8 to residue 109 of AS2 is aligned with the sequence from the corresponding region of ASL3/LBD25. The consensus sequences of the C-motif, the conserved glycine residue, and the hydrophobic residues in the leucine-zipper-like sequence are indicated by blue, red and green boxes, respectively. Amino acid residues (either similar or dissimilar) that are not identical with those in AS2 are indicated by pink characters, and the dissimilar amino acid residues are underlined. Black lines below the sequences show the C-motif (Cm), internal (In) and the leucine-zipper-like sequence (Lz) regions, respectively. (b–f) Gross morphology of various as2-1/SWAP transgenic plants. The name and the line number of each transgenic plant are indicated above the respective panel. (b) and (c) 33-day-old transgenic plants; (d), (e) and (f) 31-day-old transgenic plants. (g) Classification of the as2-1/SWAP transgenic plants with respect to complementation of the as2-1 mutation. The results of experiments with as2-1/gAS2 and as2-1/SWAP3 were obtained in Figure 4 (h) Scale bars: 10 mm.