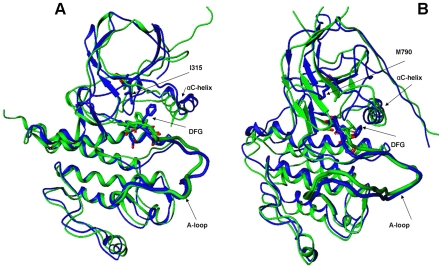
Figure 1. The predicted structural models of the ABL-T315I and EGFR-T790M mutants.
(A) Superposition of the predicted structural model of the ABL-T315I mutant (in blue) with the crystal structure of ABL-T315I (active form, pdb entry 2Z60, in green). (B) Superposition of the predicted structural model of the EGFR-T790M mutant (in blue) with the crystal structure of EGFR-T790M (active form, pdb entry 2JIT, in green). The initial ABL and EGFR structures that converged during homology modeling refinement to the crystallographic active conformations of the mutants correspond to the Src-like inactive ABL (pdb entry 2G1T) and Src-like inactive EGFR (pdb entry 2GS7).