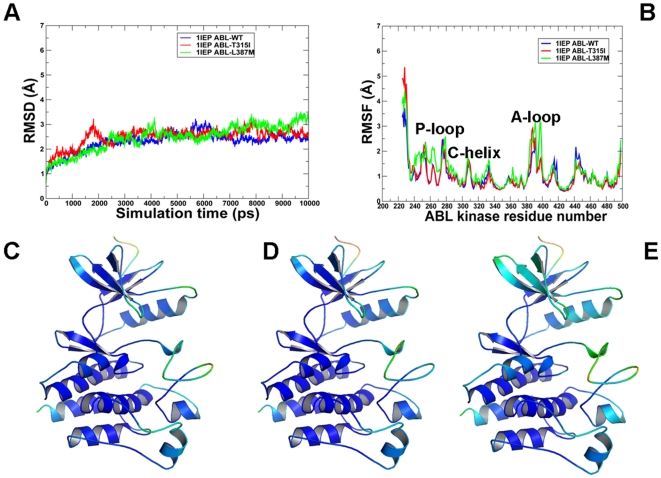
Figure 3. MD simulations of the ABL kinase domain in the imatinib-bound inactive form.
Upper Panel: (A) The RMSD fluctuations of Cα atoms and (B) the RMSF values of Cα atoms from MD simulations of ABL-WT (in blue), ABL-T315I (in red), and ABL-L387M (in green). MD simulations were performed using the Imatinib-bound, inactive form of the ABL kinase domain. Legends inside the figure panels refer to the pdb entries used in MD simulations. The color-coded sliding scheme corresponds to the following ranges of protein flexibility values: red (highly flexible with +5.00 values), brown (+4.00 values), yellow (+3.00), green (+2.00), cyan (+1.00) and blue (the least flexible). Lower Panel: Color-coded mapping of the averaged protein flexibility profiles (RMSF values) from MD simulations of the ABL-WT and ABL mutants. This mapping is presented for ABL-WT (C), ABL-T315I (D) and ABL-L387M (E). The color-coded sliding scheme corresponds to the following ranges of protein flexibility values: red (highly flexible with +5.00 values), brown (+4.00 values), yellow (+3.00), green (+2.00), cyan (+1.00) and blue (the least flexible).