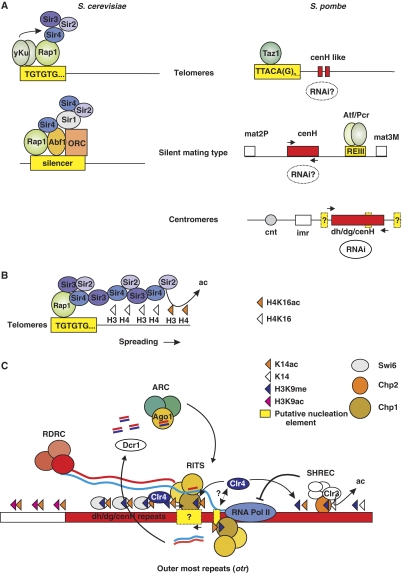
Figure 2.
Silent chromatin assembly in budding and fission yeast. (A) Cis-acting DNA sequences (nucleation sites, yellow boxes) are necessary to nucleate assembly of silent chromatin. Trans-acting proteins that directly bind the nucleation sites are indicated. Nucleation sites at fission yeast centromeres are likely to exist, although they have not been identified to date (yellow boxes). Bidirectional transcription (indicated by black arrows) of cendg/dh/H-like sequences (red boxes) is thought to produce dsRNA, which is processed into siRNAs by the RNAi machinery in S. pombe. siRNAs are required at least for the initiation of heterochromatin assembly at the silent mating-type locus and in addition for the maintenance of heterochromatin at centromeres. (B) Sir3 and Sir4 have dimerization capacity that results in the spread of the SIR complex outward from the nucleation site. Sir3 contributes to the specificity for deacetylated histone tails, whereas Sir4 enhances the affinity of the complex through its ability to bind DNA. Sir2-mediated deacetylation keeps telomeric nucleosomes hypoacetylated creating a high-affinity binding site for Sir3. (C) In S. pombe, the RITS complex promotes Clr4-mediated H3K9 methylation by associating with nascent transcripts through siRNA base pairing, and with methylated H3K9 through the chromodomain of its Chp1 subunit. Low levels of H3K9 methylation are maintained in RNAi mutant cells by a yet to be identified alternative pathway (putative nucleation element, yellow box). Primary siRNAs originating from dsRNA formed by bidirectional transcription of a centromeric sequence could prime further dsRNA synthesis and secondary siRNA generation by recruiting the RDRC complex to the nascent transcript. This would allow the spreading of H3K9me away from the nucleation site. H3K9me is bound by the chromodomain proteins Chp1, Chp2 and Swi6. The binding of Chp2 to H3K9me results in the recruitment of the SHREC complex, which in turn deacetylates H3K14. For unknown reasons this reduces RNA Pol II occupancy.