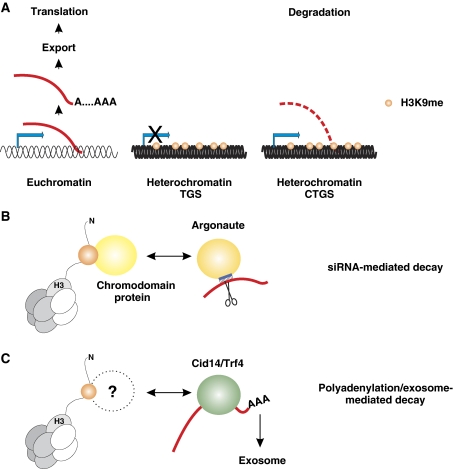
Figure 3.
Chromatin-dependent gene silencing mechanisms operate at a transcriptional and/or post-transcriptional level. (A) Silencing of heterochromatin can be achieved by either shutting off transcription (TGS) or by degradation of heterochromatic RNAs (CTGS). In contrast to classic post-transcriptional gene silencing (PTGS), CTGS depends on the status of chromatin from which the gene is transcribed and is therefore referred to as ‘co-transcriptional'. (B) RNAi-mediated degradation of heterochromatic RNAs. Argonaute-containing complexes can be physically linked to heterochromatin through chromodomain proteins. One histone-octamer is shown in grey. The chromodomain protein binds to methylated K9 (orange) of the unstructured N-terminal tail of histone H3. The siRNA (blue) guides Argonaute to the heterochromatic RNA through base-pairing interaction and induces ‘slicing'. (C) Heterochromatic gene silencing mediated by a non-canonical polyA-polymerase and the exosome. RNAs transcribed from heterochromatic regions are identified by Cid14/Trf4 and marked as aberrant with a short polyA tail. This serves as a signal for the exosome to degrade the RNA.