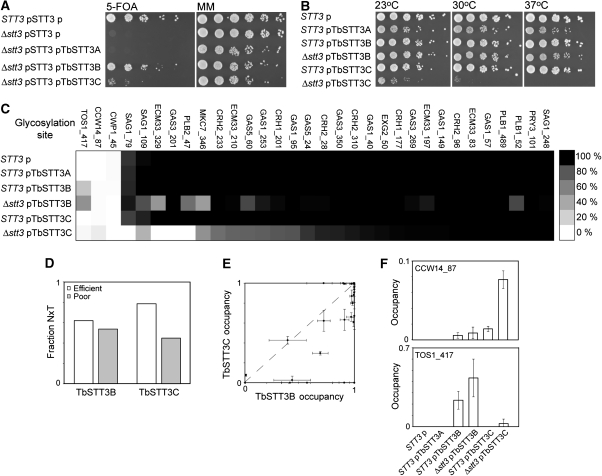
Figure 1.
Complementation of yeast Δstt3 by T. brucei OTases. (A) Yeast cells with (STT3) or without (Δstt3) genomic STT3, with the yeast STT3 gene encoded on a URA3 plasmid (pSTT3), and with various TbSTT3 genes (pTbSTT3A; pTbSTT3B; pTbSTT3C) or with empty vector (p) on a LEU2 plasmid were grown in serial dilution on minimal media (MM) or media containing 5-FOA for selection of ura-cells and (B) after selection were grown at different temperatures in serial dilution on MM. (C) Glycosylation occupancy of all sites measured for each strain, mapped from 100% occupancy, black, to 0% occupancy, white. Values are averages of three independent measurements. Data are shown in Tables SI and SII. (D) Proportion of threonine in the +2 position of efficiently and poorly glycosylated sequons. (E) Cross correlation of site-specific glycosylation efficiency by TbSTT3B and TbSTT3C. (F) Glycosylation of TOS1_417 and CCW14_87 sites by T. brucei STT3B and STT3C that are not used by yeast OTase. Error bars show range.