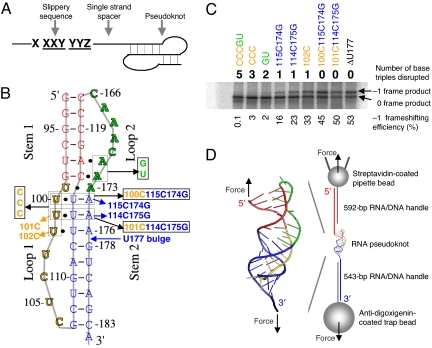
Fig. 1.
Minus-one ribosomal frameshifting (FS), pseudoknots, and experimental design. (A) Three components for programmed FS. (B) Pseudoknots used for bulk FS assay and single-molecule studies. All of the mutants were made based on ΔU177. The secondary structure of pseudoknot ΔU177 is also shown in Fig. 3C. In mutant CCCGU, all 5 base triples formed between stem 1 (shown in red) and loop 2 (shown in green) and between stem 2 (shown in blue) and loop 1 (shown in yellow) are disrupted. In TeloWT, the single nucleotide bulge U177 in stem 2 is added. (C) A typical SDS/PAGE result for the bulk FS assay. Shown on top and below the gel are the numbers of base triples disrupted and values of FS efficiency, respectively. (D) Experimental setup for mechanical (un)folding of RNA pseudoknots using optical tweezers. Pseudoknot ΔU177 is flanked by RNA/DNA hybrid handles, which are in turn attached to pipette bead and trap bead. The directions of applied force are shown with black arrows. Drawing is not to scale.