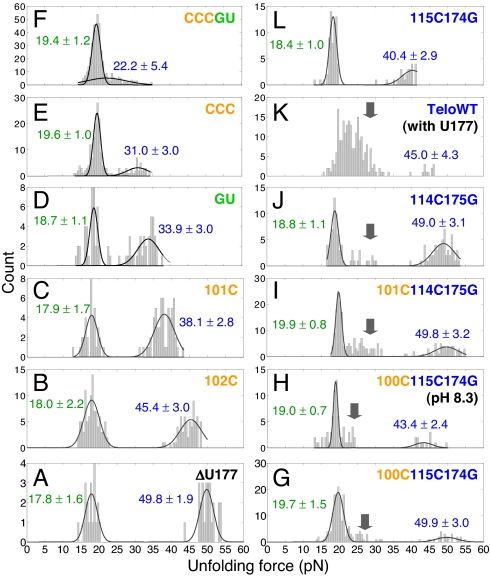
Fig. 4.
Unfolding force histograms for 1-step unfolding reactions. Values shown are peak values of Gaussian distributions with standard deviations. Mutations have significant effect on the native pseudoknots (higher unfolding forces, shown in blue) but not the folding intermediate structures (lower unfolding forces, shown in green). With the entire 5 base triples disrupted, mutant CCCGU still has 2 clusters of unfolding forces (F). Folding intermediate structures appear for pseudoknots with mutations in stem 2 (see arrows). Multiple folding intermediates may exist for TeloWT (K). All of the unfolding forces were measured at pH 7.3 except for mutant 100C115C174G, which was also measured at pH 8.3 (see H).