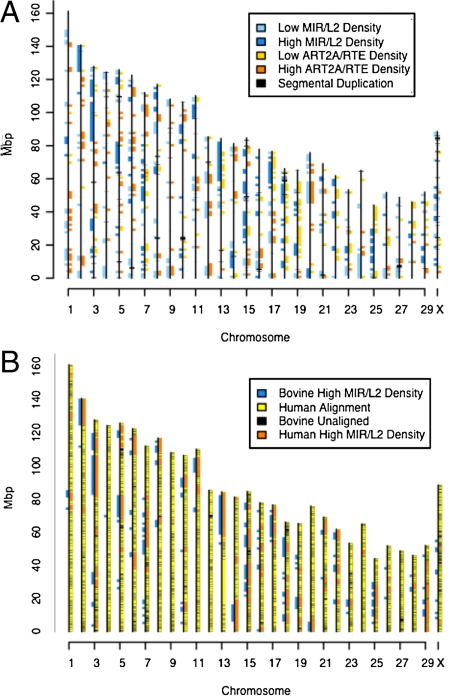
Fig. 4.
Ancestral and new repeat groups define different genomic territories. (A) Locations of 1.5-Mbp bins with extreme (high and low) ancestral (L2/MIR) and recent RTE/ART2A repeat densities are shown on the Btau4 assembly. Locations of the segmental duplications are plotted in black. Ancestral repeats tend to occur in blocks, whereas recent repeats generally do not. There is no overlap between high-density ancestral repeat blocks and bins with a high density of recent repeat blocks. Segmental duplications do not appear to colocalize with either high- or low-density regions of either repeat class. (B) Ancestral (L2/MIR) high-density bins for bovine and human are shown on the bovine assembly, along with the overall alignment from Cow-Net (23–25). Note that the “top” of the chromosomes corresponds to the end near the x axis on our plot. The y axis corresponds to nucleotide coordinates in mega base pairs (Mbp) from the bovine assembly Btau4.