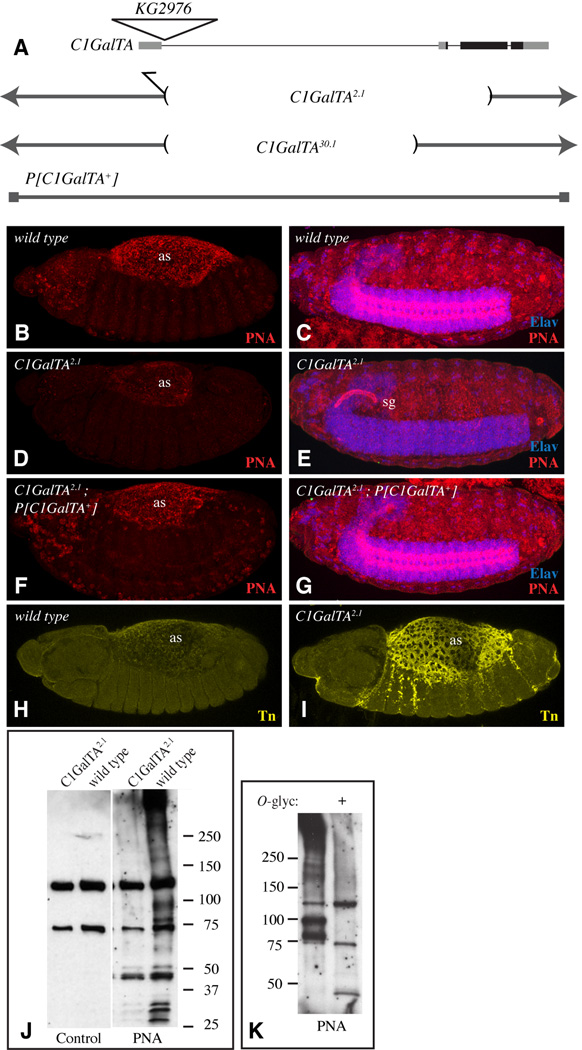
Figure 5. Influence of C1GalTA mutants on PNA and anti-Tn staining.
A) Top line shows a schematic of the C1GalTA transcription unit, with introns as thin lines, exons as thick lines, coding regions in black and non-coding regions in gray. The position of the KG2976 insertion is indicated by the triangle. Lower lines show DNA retained in mutant alleles, and DNA included in the rescue construct. B) Stage 13 wild-type embryo showing PNA staining (red) in the amnioserosa (as). C) Stage 16 wild-type embryo showing PNA staining in the CNS; the CNS is marked by anti-Elav staining (blue). D) Stage 13 C1GalTA2.1 mutant embryo showing reduced PNA staining in the amnioserosa. E) Stage 16 C1GalTA2.1 mutant embryo lacks PNA staining in the CNS, but PNA staining is now detected in the salivary gland (sg). F) Stage 13 C1GalTA2.1 mutant embryo with the P[C1GalTA+] rescue construct, PNA staining is indistinguishable from wild type. G) Stage 16 C1GalTA2.1 mutant embryo with the P[C1GalTA+] rescue construct, PNA staining is indistinguishable from wild type. H) Stage 13 wild-type embryo showing very low anti-Tn staining (yellow) in the amnioserosa. C) Stage 13 mutant embryo showing anti-Tn staining in the amnioserosa. J) PNA lectin blot on lysate from wild-type and C1GalTA2.1 mutant embryos. Two prominent bands detected in both wild-type and mutant lanes are actually background bound by the detection reagent (Avidin-hrp) as revealed by the control blot in which PNA was omitted. K) PNA lectin blot on lysate from wild-type embryos, where indicated (+) lysate was treated with O-glycanase.