Figure 4. Loss of CUL4A enhanced UV-DDB and NER activities, and reinforced the UV-responsive G1/S checkpoint to ensure genomic integrity.
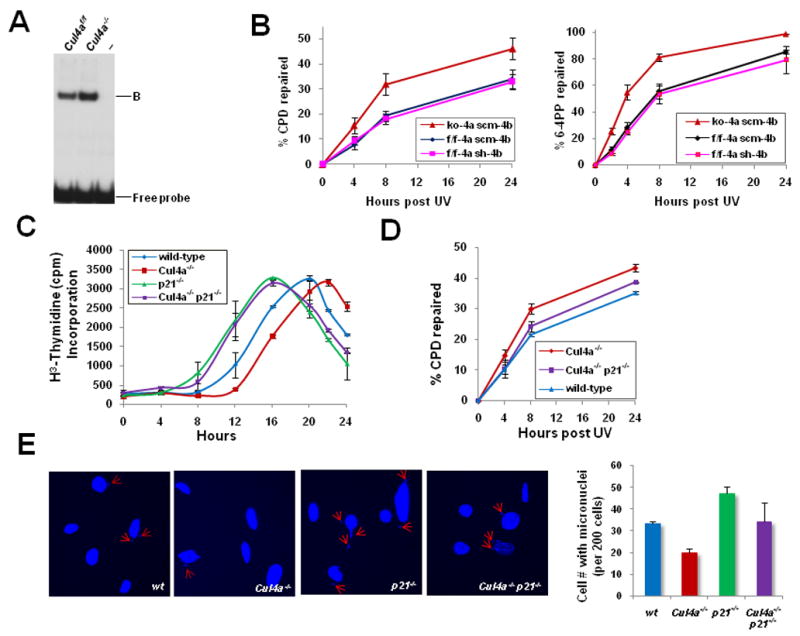
A, UV-DDB activity of Cul4af/f and Cul4a−/− MEFs was determined by electrophoretic mobility shift assay. “B”, DDB-DNA complex. B, GGR activities of CPD and 6-4PP removal were measured in primary Cul4a−/− (ko-4a scm-4b), Cul4bk/d (f/f-4a sh-4b), and control Cul4af/f (f/f-4a scm-4b) MEFs. ko, knockout; k/d, knockdown; scm, scramble. The data represent the mean and standard deviation of three experiments, with each sample point performed in triplicate. C, Primary MEFs of the indicated genotypes were synchronized, UV-irradiated and collected at the indicated time points for [3H]-thymidine incorporation assay. D, GGR for removal of CPDs was measured in primary wild-type, Cul4a−/−, and Cul4a−/− p21−/− MEFs as described in Fig. 4C. E, Micronuclei formation in wild-type, Cul4a−/−, p21−/− and Cul4a−/− p21−/− MEFs, as measured by staining with DAPI at 48 hours post-UV. Error bars represent standard deviations of 3 independent experiments.
