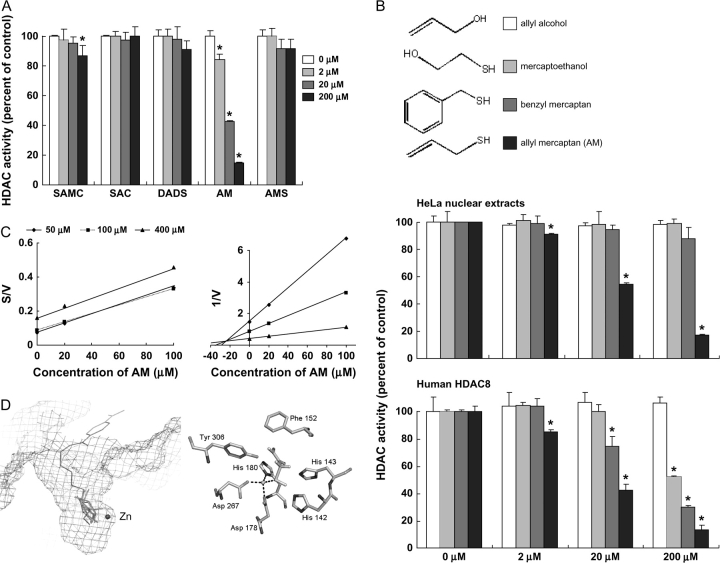
Fig. 1.
AM is a competitive HDAC inhibitor. (A) HDAC activity was evaluated using HeLa nuclear extracts in the presence of 2, 20 and 200 μM SAMC, SAC, DADS, AM and allyl methyl sulfide (AMS). Data (mean ± SD, n = 3, *P < 0.05) were expressed as percent of dimethylsulfoxide control. (B) Inhibition of HDAC activity by AM and three structural analogues, allyl alcohol, benzyl mercaptan and mercaptoethanol. HDAC activity was assayed with HeLa nuclear extracts (top) or human HDAC8 (bottom). Data = mean ± SD, n = 3, *P < 0.05. (C) Cornish-Bowden plot (left) and Dixon plot (right) of HDAC8 inhibition by AM, indicating competitive binding (Ki = 24 μM). (D) Modeling of the HDAC8–AM complex using MacroModel® v8.5 (Schrödinger). Left: lowest energy configurations of AM in the active site of human HDAC8 based on a non-bonded model docking search. Enzyme-bound inhibitor MS-344 (brown) and the active zinc atom (blue sphere) are shown from a prior report (37), which highlights the favorable orientation of AM (green). Right: AM docked in the HDAC8 catalytic core in the lowest energy structure after Jaguar calculation (Schrödinger). The sulfur of AM (yellow) was oriented 2.25 Å from the zinc atom (blue sphere), and hydrophobic interactions with adjacent residues were predicted to further stabilize AM binding in the HDAC8 pocket.