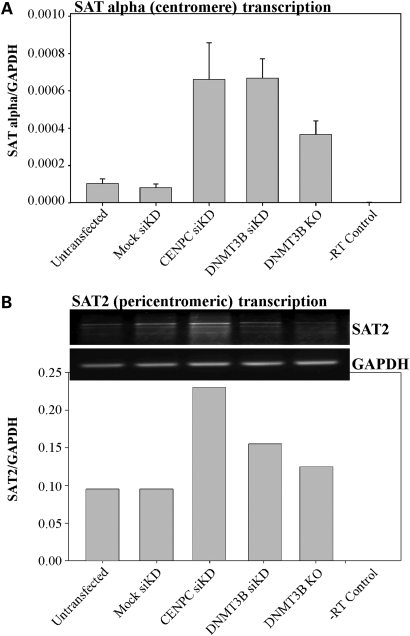
Figure 8.
Loss of CENP-C and DNMT3B-mediated epigenetic marks at the centromere region result in elevated levels of repeat transcription. (A) Quantitative RT–PCR analysis of alpha satellite repeat transcripts in HCT116 cells either mock transfected or transfected with siRNA targeting CENP-C or DNMT3B. HCT116 cells with a genetic knockout of DNMT3B (3BKO) are also shown. Values are the average of triplicate RT–PCR reactions relative to GAPDH as a loading control. The error bar is the standard deviation from the mean. (B) Semi-quantitative RT–PCR analysis of satellite 2 repeat transcription from the same knock down/knockout panel as in (A). RT–PCR reactions were repeated three times and a representative ethidium bromide stained agarose gel photo is shown. Bands were quantified using BioRad Quantity One software and set relative to an independent amplification for GAPDH (lower panel). Although samples were DNase treated, we included a no RT (-RT) control to ensure that we were not amplifying contaminating satellite DNA in the RNA preparation.