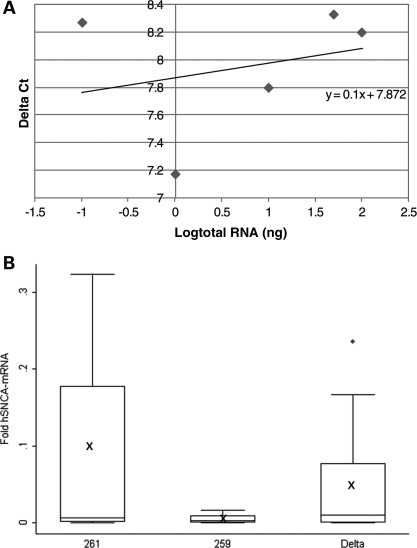
Figure 6.
Effect of the SNCA-Rep1 promoter genotypes on human SNCA-mRNA expression levels in transgenic mouse whole blood. (A) Validation curve of the Δ real-time assay for relative quantization of human SNCA-mRNA relative to mouse Gapdh-mRNA in blood. Relative efficiency plots of human-SNCA and mouse-Gapdh were formed by plotting the log input amount (ng of total RNA) versus the ΔCt = [Ct(SNCA)−Ct(Gapdh)]. The slope is 0.1, which indicated the validation of the ΔCt calculation in the range of 0.1–100 ng RNA. (B) Fold levels of human SNCA-mRNA were assayed by real-time RT–PCR and calculated relative to mouse Gapdh-mRNA reference control using the 2−ΔCt method. No detectable products of the human SNCA-mRNA reaction were observed when WT mouse blood-cDNA was used as template. The risk genotype, 261/261, correlates with higher SNCA-mRNA levels than the protective genotype, 259/259. The deleted SNCA-Rep1 genotype shows no significant correlation with SNCA-mRNA levels in comparison with genotypes 261/261 and 259/259 (P = 0.524). In total, 72 mice were analyzed; for each genotype (261/261, 259/259 and Δ/Δ), the box plot represents the analysis performed using two transgenic lines, 12 animals from each line (6 males and 6 females); overall, 24 whole-blood samples (per genotype) were each repeated twice, independently. The average values are presented by ‘X’. The box plot shows the median (horizontal line inside the box) and the 25th and 75th percentiles (horizontal borders of the box). The range between the 25th and 75th percentiles is the interquartile range. The whiskers show the minimal and maximal values inside the main data body.