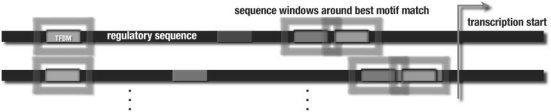
Fig. 1.
The idea behind the RM kernel: a motif finder is applied to the regulatory sequences in the input set (long, dark bars), which identifies overrepresented motifs (short, light bars). The best matching motifs (boxed) in every sequence serve as starting points, where we excise a window of 20 bp around the center of each motif occurrence for the WDSC kernel. Conservation information for these windows is looked up in a precomputed multiple genome alignment (cf. Section S.2 of the Supplementary Material for details on conservation data). Additionally, we construct an input vector for the RBF kernel of the pairwise motif distance, and distance to the transcription start (if available).