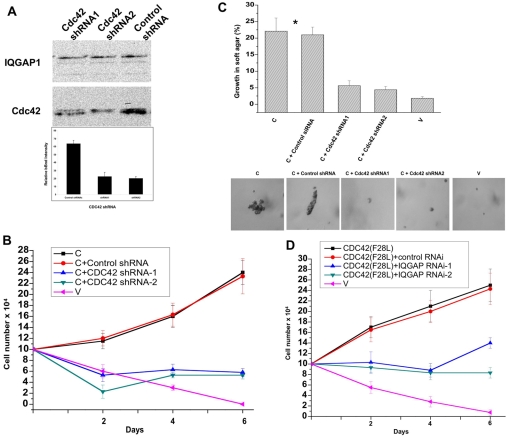
Fig. 5.
Mutual requirement of IQGAP1 and CDC42 for cellular transformation. (A) Upper panel, western blot of transformed IQGAP1-C cells that were treated with two shRNAs targeting CDC42 and evaluated by blotting with antibodies for CDC42 and for IQGAP1 as a control. Lower panel, densitometry quantification of the CDC42 band in control (first column) and shRNA-treated (second and third columns) bands demonstrating ∼33% average knockdown of endogenous CDC42 by shRNA treatment. (B) Growth in low-serum medium of IQGAP1-C and control (V) NIH3T3 stable cells untreated or treated with CDC42 shRNA or control shRNA. (C) Growth in soft agar of the same cell lines. Upper panel, quantification of colonies greater than 50 μm in diameter. Means ± s.e.m. for n=3 experiments are shown for four clones done in duplicate. Growth of IQGAP1-C cells with and without control siRNA was significantly greater than for control cells (*P<0.001). Lower panels, representative photomicrographs of colonies formed by the different cell lines on day 10. (D) Growth in low-serum medium of transformed CDC42F28L and vector-control NIH3T3 cells untreated or treated with specific IQGAP1-siRNA.