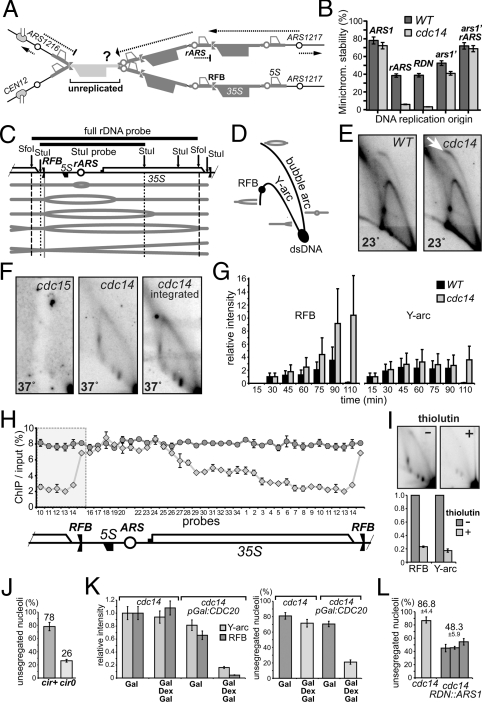
Fig. 1.
Nucleolar missegregation in cdc14 is consistent with rDNA under-replication. (A) Putative linking of rDNA sister chromatids due to under-replication, leading to the segregation failure. Only three repeats are shown for simplicity. Dotted lines - paths of replication forks. (B) rARS cannot maintain minichromosomes in cdc14 cells. The minichromosome stability was tested in isogenic strains under selective conditions at 23 °C. TRP1/CEN3 minichromosomes had various replication origins: full-length rDNA, short rARS-containing fragments, full-length ARS1 and a truncated (weakened) ARS1 (ars1′). Minichromosomes containing rARS (pAS1071) or full-length rDNA repeat (pAS1072) as a single replicator failed to propagate in cdc14 cells: all surviving Trp+ clones contained TRP1 integration. The rARS is not completely inactive in cdc14-1 cells, as indicated by the synergistic restoration of minichromosome stability when ars1′ and rARS are combined (last column). (C) Schematic of a single rDNA repeat progressing through replication stages. Southern hybridization probes and restriction sites used for 2D gel analyses (SfoI or StuI) are shown. (D) Replication intermediates distribution on a neutral-neutral two-dimensional gel. (E) Replication intermediates in asynchronous wild type and cdc14 mutant at permissive temperature. Wild type and cdc14-1 cells were grown at 23 °C. Purified DNA was digested with SfoI, enriched by ssDNA affinity chromatography, separated by 2D electrophoresis and analyzed by Southern blotting. The arrow: relative enrichment of the bubble arc. (F) cdc14 mutants arrested at 37 °C still contain rDNA replication intermediates. Cultures were arrested in G1 for 2 h at 23 °C, 1 h at 37 °C, and released at 37 °C for 3 h. Anaphase-arrested cells were analyzed as in E, except that rDNA was digested with StuI. To exclude the possibility that the observed retarded replication is a property of a hidden (non-CDC14) mutation, the cdc14-1 allele was cloned by PCR and reintroduced it into a wild-type strain by gene replacement (cdc14 integrated). (G) The cdc14 mutation delays progression of rDNA replication. Cultures were synchronously released from G1 arrest as in F, time-course cell samples were collected every 15 min and analyzed as in E. RFB and Y-arc signals were normalized to the 30 min and 15 min points, respectively, as signals at these time points were similar for wild type and cdc14. (H) BrdU incorporation in rDNA is progressively inhibited in cdc14 cells in the RNA PolI-transcribed region. Both strains were arrested as in G and BrdU was added to the media 30 min before the release from G1 into fresh YPD containing BrdU. BrdU incorporation was analyzed 2 h after release by ChIP/qPCR using rDNA probes as in (14). Shaded area is duplicated to show RFB surroundings. (I) Inhibition of transcription reduces underreplication in cdc14. The cdc14-1 mutant was arrested and released as in F in the absence or presence of thiolutin (87 μM), which was added in the mid-S-phase. The RFB and Y-arc signals were quantified and normalized to the corresponding values without thiolutin. (J) Curing cdc14 strains of 2-μm episomes significantly rescues nucleolar missegregation. Four independent cir0 clones were analyzed after arresting cells as in F. Minimum 200 nuclei were scored. (K) Metaphase delay suppresses both the rDNA replication delay and nucleolar segregation defects in cdc14 mutants. pGal:CDC20 and control cells were arrested in YPRG media in G1 and then arrested in metaphase (the Cdc20p depletion after addition of dextrose) with inactivated Cdc14p (37 °C). Cultures were grown in YPRG and split in half after the G1 arrest, one-half was released to YPRG at 37 °C for 5 h (Gal), while another half was released to YPD at 37 °C (to arrest cells in metaphase). Cells were harvested after 3 h, re-suspended in prewarmed YPRG media and incubated at 37 °C for 2 h (Gal Dex Gal). Continuous CDC20 expression (Gal) was used to control for possible overexpression effects unrelated to the mitotic arrest itself. Replication intermediates were analyzed as in E; the nucleolus segregation was scored by Sik1p-RFP, in 200 anaphase cells. (L) ARS1 integration in the rDNA locus partially rescues nucleolar nondisjunction in cdc14. Three control cdc14 cultures and three independent clones with ARS1 integration into rDNA were grown at 23 °C and arrested at 37 °C for 3 h. Cells were fixed and stained with DAPI, nucleolar segregation was monitored by Sik1p-RFP, with two sets of 100 cells scored per each strain.