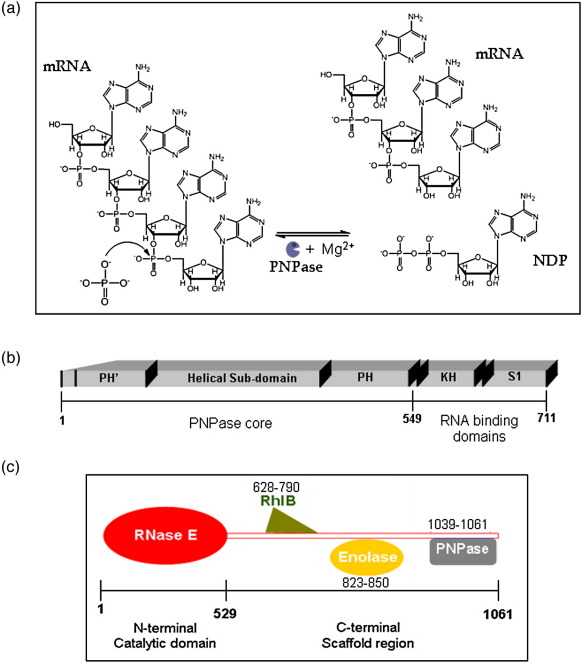
Fig. 1.
Polynucleotide phosphorylase catalytic activity, domain organisation and interaction site in the RNA degradosome assembly. (a) Phosphorolytic and polymerisation activities of PNPase. Adenine is shown for illustration, but the enzyme can use any RNA base. (b) E. coli PNPase domain structure. The key domains of PNPase and the boundaries for the “core” used in this study are shown. (c) Schematic of the canonical degradosome protomer, including the site of interactions of the degradosome components with the C-terminal scaffold region of RNase E. Included in the cartoon are the interaction sites for the DEAD-box helicase RhlB (green), polynucleotide phosphorylase (PNPase, grey), and enolase (yellow). Two RNA-binding sites flank the RhlB interaction region (not shown).