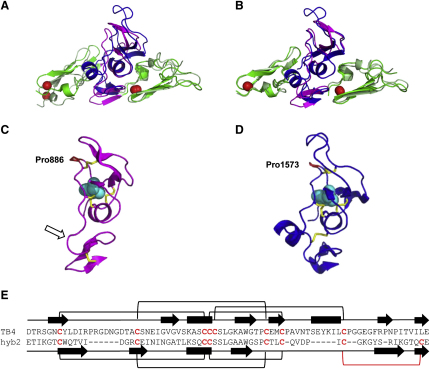
Figure 3.
Comparison of Hyb2 and TB4 Domain Structures
(A) Superposition of cbEGF9-hyb2-cbEGF10 onto the previously determined structure of fibrillin-1 fragment cbEGF22-TB4_cbEGF23 (Lee et al., 2004) showing the overall similarity of the structures.
(B) Superpostion of individual domains of cbEGF9-hyb2-cbEGF10 onto the corresponding regions of cbEGF22-TB4-cbEGF23 indicated that differences in the structures were largely due to small differences seen in the interdomain packing interactions of the different constructs.
(C and D) Domain hyb2 (C) has secondary structure elements similar to those seen in TB4 (D) (Lee et al., 2004). A conserved tryptophan (cyan spheres in the 3D models) is involved in forming the hydrophobic core of each domain. The arrow in (C) highlights the loss of an α helix in hyb2 relative to the corresponding position in TB4.
(E) Comparison of the TB4 and hyb2 domain sequences with the disulphide bond pairings in each indicated. The rearrangement of disulphides in hyb2 compared with TB4 results in the stabilization of a C-terminal β sheet in the hyb2 domain (red bracket).