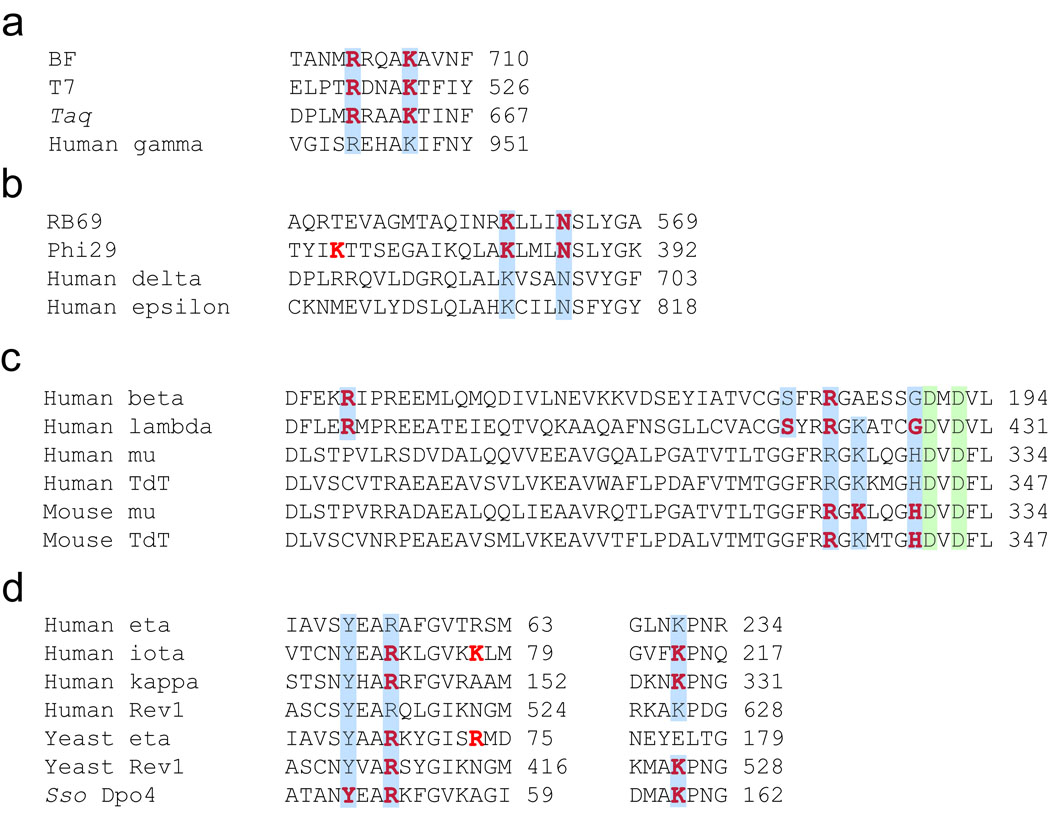
Figure 9.
Conservation of positively charged residues involved in stabilizing the triphosphate moiety and/or the pyrophosphate product. Based upon X-ray crystal structural analysis, amino acid residues within 4.0 Å of the triphosphate moiety (red) were identified. Amino acid sequences of selected (A) A-family, (B) B-family, (C) X-family, and (D) Y-family DNA polymerases were aligned using ClustalW2. Conserved residues are shaded in blue and the conserved catalytic aspartic acid residues are shaded in green.