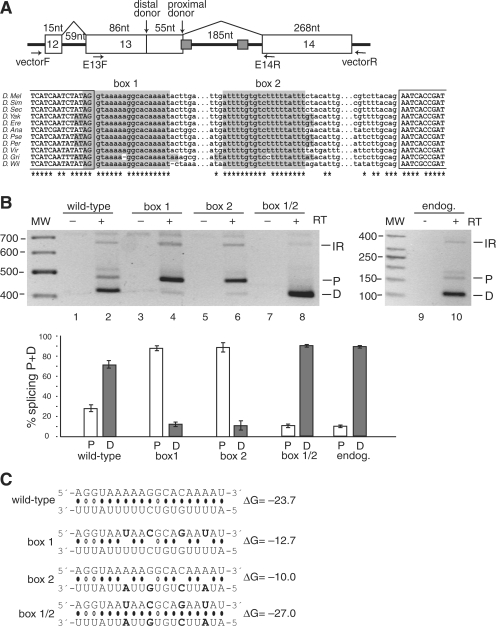
Figure 2.
Splicing to alternative donor sites in the CG33298 mini-gene is regulated by the stem structure formed by the conserved box sequences. (A) Top panel: Schematic representation of the CG33298 mini-gene, which contains the chromosomal region 2L:9519321–9519987. The box 1 sequence overlaps with the proximal donor splice site of exon 13. The location of the primers used for PCR amplification of the mini-gene mRNAs (in the vector) and the endogenous mRNAs (in the exons) are indicated. Bottom panel: Multiple sequence alignment for the intronic regions containing the boxes. 100% conserved positions are indicated by asterisks. No orthologous sequence was found for D. mojavensis. Complementary boxes (highlighted) are conserved in all but two positions; the conservation rate for the rest of the intron is less than 3%. (B) Secondary structure formed by the conserved boxes affects donor site usage. mRNA products from either the mini-gene (lanes 1–8) or from the endogenous gene (lanes 9–10) were analyzed by RT–PCR. Bands are labeled as D, distal donor; P, proximal donor; or IR, intron 13 retention. The addition (+) or absence (−) of the reverse transcriptase (RT) enzyme to the reaction is indicated. The results of three independent splicing assays are represented graphically in the bottom panel, as the ratio of the band intensity (P or D) to the total intensity (P + D). (C) Predicted base-pairing for the wild-type, box 1, box 2 and box 1/2 mutants (point mutations are shown in boldface), with the estimated equilibrium free energies (given in kcal/mol). The box 1 sequence is shown above the box 2 sequence.