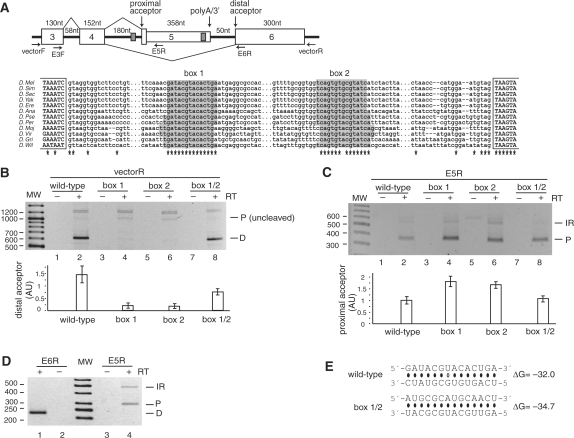
Figure 4.
A stem structure regulates alternative usage of acceptor splice sites in the Nmnat (CG13645) mini-gene. (A) Top panel: Representation of the Nmnat mini-gene (chromosome 3R:20771699–20772905). Exon 5 is an internal terminal exon that is cleaved when included (the poly-adenylation/3′-processing signals are indicated), while exon 6 is an internal exon. Box 2 is located upstream of the poly(A) signal in exon 5. Primers used in the PCR amplifications are indicated. Bottom panel: Multiple sequence alignment of the sequence between exons 4 and 6. The complementary boxes are almost 100% conserved, with only one change of GU to AU in the base pairs. (B) Splicing products from the mini-gene were amplified with a reverse primer to the vector to amplify the isoforms formed by splicing to the distal acceptor (D) or to the proximal acceptor (P) that had not been cleaved. The results of three independent splicing assays are represented graphically in the bottom panel for distal acceptor usage. Samples were normalized prior to loading against an independent PCR performed in parallel with a reverse primer to exon 4, to visualize the constitutively-spliced product of exon 3–exon 4 (data not shown). (C) As in (B), except that a reverse primer in exon 5 was used to amplify splice products to proximal acceptor (P) or with intron 4 retention (IR). Proximal acceptor usage is depicted graphically at the bottom. (D) Endogenous mRNA was amplified with reverse primers in exon 6 or exon 5. (E) Predicted base pairing for the wild-type, box 1, box 2 and box 1/2 mutants (point mutations are shown in boldface), and their estimated equilibrium free energies. Since the sequence was completely exchanged during mutagenesis, no base-pairing is predicted to occur for the single box mutations (box 1 and box 2).