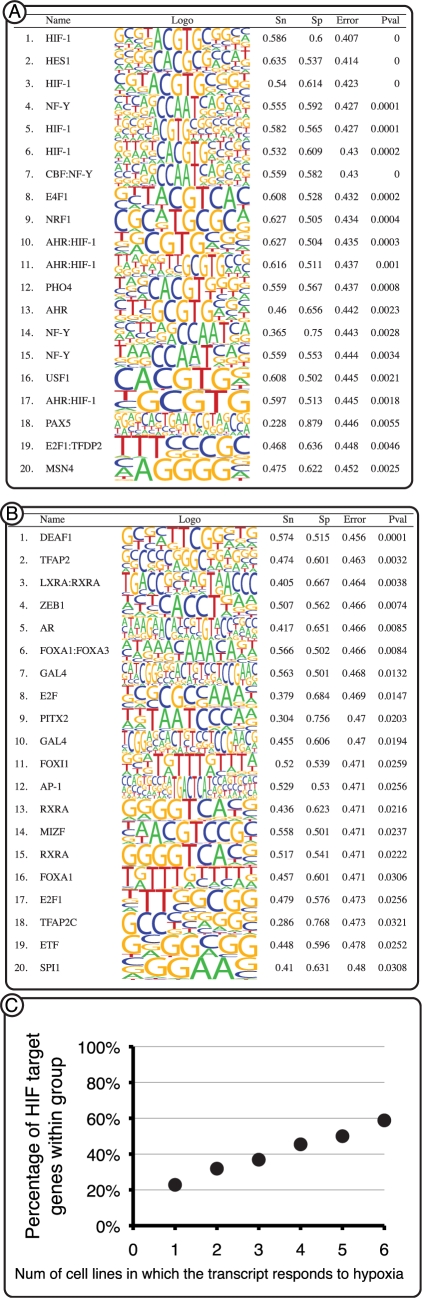
Figure 2.
(A and B) Binding site enrichment analysis. The program MOTIFCLASS was employed to identify binding sites significantly over-represented in the promoters of genes that respond to hypoxia in B cells. (A) Totally 770 promoters of genes that responded to hypoxia in which a detectable HIF-1-binding site was identified. HIF-1 PWMs were identified as the most enriched. (B) Totally 1456 promoters of genes that responded to hypoxia but do not contain a detectable HIF-1-binding site. No HIF-1 PWM was identified in the top 20 matrices. Sn, sensitivity; Sp, specificity; Error, classification error rate; Pval, P-value. (C) Correlation between the number of HIF-target genes and the number of cells in which they responded to hypoxia. Only 31% of the genes that responded to hypoxia in one of six cells was predicted as a HIF-target gene, compared to 71% of the genes that responded to hypoxia in six of six cell lines.