Abstract
Retroviral insertional mutagenesis has been instrumental for the identification of genes important in cancer development. The molecular mechanisms involved in retroviral-mediated activation of proto-oncogenes influence the distribution of insertions within specific regions during tumorigenesis and hence may point to novel gene structures. From a retroviral tagging screen on tumors of 1767 SL3-3 MLV-infected BALB/c mice, intron 2 of the AP-1 repressor Jdp2 locus was found frequently targeted by proviruses resulting in upregulation of non-canonical RNA subspecies. We identified several promoter regions within 1000 bp upstream of exon 3 that allowed for the production of Jdp2 protein isoforms lacking the histone acetylase inhibitory domain INHAT present in canonical Jdp2. The novel Jdp2 isoforms localized to the nucleus and over-expression in murine fibroblast cells induced cell death similar to canonic Jdp2. When expressed in the context of oncogenic NRAS both full length Jdp2 and the shorter isoforms increased anchorage-independent growth. Our results demonstrate a biological function of Jdp2 lacking the INHAT domain and suggest a post-genomic application for the use of retroviral tagging data in identifying new gene products with a potential role in tumorigenesis.
INTRODUCTION
The cJun dimerization protein 2 (Jdp2) gene encodes a 163-amino-acid protein, Jdp2, initially discovered as an inhibitor of AP-1/TRE (TPA response element)- and AP-1/CRE (cyclic AMP response element)-mediated transcription through heterodimerization with Jun and ATF2, respectively (1,2). Additionally, Jdp2 may form homodimers and heterodimerize with JunD, JunB, the CCAAT/enhancer-binding protein C/EBPγ and C/EBP homologous protein (CHOP) (1,3–5). Jdp2 has been shown to inhibit p300/ATF2-mediated transactivation of cJun upon retinoic-acid-induced commitment of murine F9 cells by recruiting histone deacetylase 3 (HDAC3) to the promoter of cJun (6), and to inhibit the histone acetyltransferase activity (7). This inhibition of histone acetyltransferase (INHAT) activity is associated with the N-terminal domain encoded by exon 2. In addition to acting as a transcriptional repressor, Jdp2 also directly associates with the progesterone receptor (PR) and potentiates ligand-dependent PR-mediated transactivation (8). Furthermore, Jdp2 in a complex with CHOP10 was recently shown to strongly enhance TRE but not CRE-dependent transcription (5).
Jdp2 is involved in diverse processes. Over-expression in vitro leads to differentiation of osteoclast (9) and myoblast (10) cell lines, and recent information from knock-out mice has shown that Jdp2 works as a repressor of adipocyte differentiation (11). The underlying mechanism was shown to involve the inhibition of histone H3 acetylation of the promoter of the adipogenesis-related gene C/EBPδ (11). In other settings, Jdp2 appears to be involved in the inhibition of apoptosis (12,13), as well as in p16Ink4a-mediated induction of replicative senescence (14). Finally, general inhibition of the AP-1 complex by expression of Jdp2 specifically in the heart correlates with the induction of atrial dilatation (15).
Depending on the context, Jdp2 displays both oncogenic (16–20) and tumor-suppressive (10,21) properties. The regulation of Jdp2 is poorly understood. At the RNA level, a ubiquitous expression pattern involving several mRNA species is observed (2,18). Furthermore, upon exogenic insults the C-terminus of the Jdp2 protein is specifically phosphorylated by the c-Jun N-terminal kinase (JNK) (22,23).
The murine leukemia virus (MLV) is a non-oncogene bearing retrovirus, which induces lymphomas in susceptible mice by insertional mutagenesis of host genes. Assuming random integration, superimposing retroviral insertion sites from a cohort of infected mice reveals common integration sites (CIS), loci defined as having significantly more insertions than would be expected by chance (24). CISs point to potential cancer-associated genes that have been deregulated by the integrated retrovirus. Various modes of activation have been proposed according to the position and transcriptional orientation of the virus relative to the activated gene. Two commonly observed activation mechanisms are enhancer and promoter activation (25). In the latter case, transcription initiates from the provirus promoter resulting in a virus-host fusion mRNA. Enhancer activation refers to positive influence from cis-elements in the proviral enhancer on the promoter of the host gene. Enhancer activation may occur over large distances, and for unknown reasons there appears to be a selection for the opposite (integration upstream of the host gene) or the same (integration downstream of host gene) transcriptional orientation of provirus as compared to host gene.
We here describe the usage of integration distribution patterns in Jdp2 to identify novel transcriptional structures in the second intron of the Jdp2 gene resulting in the activation of Jdp2 protein isoforms. Using assays for cell survival and colony formation, we demonstrate a functional activity of these novel isoforms that lack the N-terminal INHAT domain. Our results suggest a general approach of using retroviral tagging data to identify transcriptional structures and possible protein isoforms of genes involved in cancer development.
MATERIALS AND METHODS
Identification of integration sites
Newborn BALB/c mice were infected with SL3-3 MLV and treated as described previously (26). On sickness or observation of tumors, mice were sacrificed, and splenic and thymic tumors removed and stored at −80°C. Genomic DNA from tumors was isolated using DNeasy Tissue Kit (Qiagen) and provirus integration sites were identified using the Splinkerette-based PCR method as described (26).
PCR screening of the 3′ end of Jdp2 intron 2 for integrations was done on 0.05–0.5 μg genomic tumor DNA using recombinant Taq DNA polymerase (Invitrogen) following the manufacturer's recommendations, and using virus LTR-specific primers (either LTR-2620, 5′-gaattcgatatcgatccccggtcatctggg-3′ or LTR-440, 5′-ttcataaggcttagccagctaactgcagt-3′) together with a reverse exon 3-specific primer Exon3-52, 5′-ttcttcttgttccggcatct-3′.
RT-PCR and quantitative real-time PCR
Total RNA was extracted from tumor samples using TRIZOL (Invitrogen) and stored immediately at −80°C until use. The integrity of the isolated RNA was assessed by visual inspection of the intensity of the 28S and 18S ribosomal bands on a non-denaturating ethidium-bromide-stained agarose gel. RNA quantity was determined spectrophotometrically. Two μg total RNA was reverse transcribed using the First Strand cDNA Synthesis Kit (Fermentas) following the manufacturer's recommendation with the provided poly(dT) oligo. To look for alternative splicing between published exons 1a through 1d, PCR reactions were done with forward primers Exon1a-154 (5′-tgggcaccgcgcctgcagcag-3′), Exon1b-148 (5′-ggaggagcgcgagcat-3′), Exon1c-70 (5′-gctctggctgggttaggagggaac-3′) or Exon1d-150 (5′-cagctgcctctctccatctt-3′) and reverse primer Intron2-87 (5′-tccttcgctcttcttcctcgtctagctt-3′) using 1/500 of the cDNA (corresponding to 4 ng of total RNA) per PCR reaction (Taq polymerase, Invitrogen). Each PCR reaction consisted of an initial melting step at 95°C for 5 min followed by 35 cycles of amplification, each consisting of a 30 s melting step at 95°C, annealing at 55–60°C for 30 s and elongation at 72°C for 45 s. A final elongation step at 72°C was done for 10 min. Quantitative real-time PCR (QRT-PCR) was done using Platinum SYBR Green QRT-PCR SuperMix UDG (Invitrogen) following the manufacturer's recommendations. Reactions in Figures 5 and 6 were run in triplicates at least two times on a Mx3005 apparatus (Stratagene) with cDNA corresponding to 10 ng of total RNA, whereas the initial screen in Figure 2 was done two times in duplicate. Relative quantification was done with the Window-of-Linearity-based method applying LinReqPCR software (version 11) using an averaged amplification efficiency per amplicon per run (27). We estimated an approximate copy number (absolute quantification) by relating threshold cycles of unknown samples to a standard curve made from amplicon-containing plasmid dilutions. The signal was normalized to the expression level of the TATA box binding protein (Tbp) or the geometric mean of Tbp and Actb (β-Actin) (28). Primers in Jdp2 were: Exon2-51, 5′-gaggtgaaactgggcaagag-3′; Exon3-52, 5′-ttcttcttgttccggcatct-3′; Intron2-55, 5′-tttgctgagatgtgggtgag-3′; Exon4-57, 5′-gcggttgagcatcaggataa-3′; E1e-161, 5′-ggtgtctaatcggtggctct-3′; E1f-164, 5′-tgtgttgaaaggatctgtcaagct-3′ (underscored base pairs are specific to the first bases in exon 3 to guarantee specificity to exon 1f–exon3 mRNA; this was ensured by the inability of amplification of plasmid-containing exon 1e under the applied PCR conditions (data not shown)). Primers in Tbp and Actb were Tbp-60, 5′-agagagccacggacaactg-3′; Tbp-61, 5′-actctagcatattttcttgctgct-3′; Actin-165, 5′-acacagtgctgtctggtggt-3′; Actin-166, 5′-ctggaaggtggacagtgagg-3′. PCR conditions consisted of an initial melting step of 95°C for 10 min, and then amplification was conducted through 40 repetitions of a 95°C step for 15 s and a combined annealing and extension step at 60°C for 30 s. In case of amplicon E1f-E4, the annealing/extension step was at 62°C. Lack of unspecific amplification was ensured with a final melting curve analysis from 55°C to 95°C.
Figure 5.
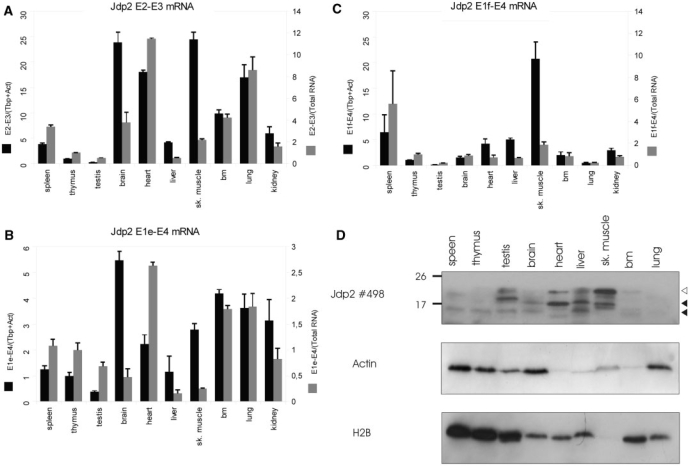
Jdp2 isoforms are differentially expressed in the normal tissue. (A–C) QRT-PCR was done in triplicates as described for Figure 5 on the indicated BALB/c mouse tissues to detect full length (A), exon 1e-3-4 (B) and exon 1f-3-4 (C) mRNA. Expression signal is shown as normalized to the geometric mean of Actb and Tbp (black bars) or normalized to total RNA (white bars) and is shown as fold difference to thymus. The figures are representative of two–three experiments. (D) Western blotting on 0.2 μM PVDF membranes using polyclonal anti-Jdp2 and, subsequently, anti-β-Actin and anti-H2B antibody on crude protein extracts from the same panel of BALB/c tissue. Open and closed arrows indicate the position of full length and isoform Jdp2.
Figure 6.
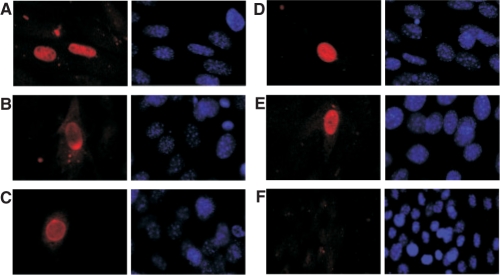
Jdp2 isoforms localize to the nucleus. NIH3T3 cells were transiently transfected with vectors expressing N-terminally FLAG tagged full length Jdp2 (A) or either of three N-terminally Jdp2 isoforms, Jdp2-132 (B-C), Jdp2-119 (D) or Jdp2-107 (E). As controls are shown cells transfected with empty vector (F). The subcellular localization was revealed by immunocyotchemistry using an anti-FLAG primary antibody (left). Cells were co-stained with DAPI dye (right). The results are representative of three independent experiments.
Figure 2.
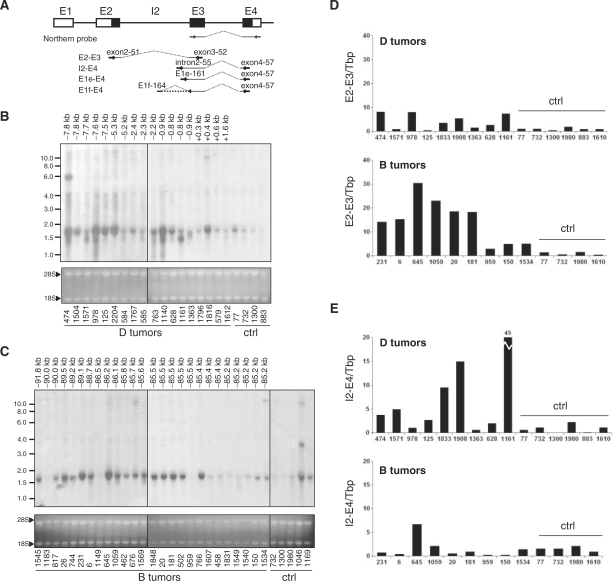
Correlation of intragenic provirus insertion and appearance of Jdp2 intron 2 including transcripts. (A) Schematic representation of the localization of the Northern blot probe and QRT-PCR amplicons E2-E3, E2-E4, I2-E4, E1e-E4 and E1f-E4 for Jdp2 mRNA detection with exons shown as boxes and coding sequence in black. (B and C) Northern blotting on total RNA from thymus tumors from a subset of animals with integration in clusters D (B) and B (C) according to the retroviral tagging data. Control samples (Ctrl) are thymic tumors from retrovirus-infected animals of the same cohort in which no integrations were found by retroviral tagging. The distance between integration and the beginning of Jdp2 exon 3 for each tumor is shown above the lanes. Ethidium bromide staining of ribosomal bands 28S and 18S was used to evaluate even loading and RNA integrity. (D and E) QRT-PCR on a subset of D tumors and B tumors amplifying either Refseq Jdp2 mRNA (E2-E3) (D) or intron 2-including alternative mRNA (I2-E4) (E). The signal was normalized to Tbp and shown as fold difference to the average of control tumor samples (Ctrl).
5′ RACE
5′ Rapid amplification of cDNA ends (5′ RACE) was done utilizing the SMART™ RACE kit (Clontech) following the manufacturer's recommendations. One μg total RNA from tumor 1161, uninfected BALB/c bone marrow and brain were reverse transcribed with PowerScript™ Reverse Transcriptase using an oligo(dT) primer during each SMART RACE reaction. 5′ RACE PCR was subsequently done on 1/120 of the cDNA (corresponding to 8 ng total RNA) with the PCR conditions: 5 min initial melting step at 95°C, then 6 cycles of annealing step-touch-down from 74 to 68°C followed by 30 cycles of amplification with annealing temperature at 67°C and a final 10 min elongation step. Each amplification cycle consisted of a 30 s melting step, 30 s annealing step and 30 s elongation at 72°C. Primers were a mixture of linker-specific (UPM) forward primers (comes with the kit) and either of the gene-specific reverse primers Exon3-96 (5′-gcatctggctgcagcgactttgt-3′), Exon3-46 (5′-gcagaaactctgtgcgttccttcttc-3′) or intron2-86 (5′-cgattagacaccatgggtggaactgagc-3′). The transcription start site (TSS) distribution in tumor tissue was similar when using either exon 3-specific reverse primer, and hence the TSS data were pooled. A control reaction was carried out with a reverse primer in Actb (Actin-104, 5′-ctggaaggtggacagtgagg-3′). With regard to bone marrow and brain, a nested PCR was done subsequently with forward and reverse primers NUP (comes with the kit) and Exon3-97 (5′-gctgcagcgactttgttctt-3′), respectively. Of each 50 μl 5′ RACE PCR, 2 μl was cloned into pCR4 using the TOPO TA cloning kit (Invitrogen) and transformed into One Shot TOP10 competent cells (Invitrogen). Bacteria were selected on ampicillin and screened for insert using TEMPase Hot Start DNA Polymerase (Ampliqon) with vector primers M13-110 5′-gtaaaacgacggccag-3′ and M13-111 5′-caggaaacagctatgac-3′. The same primers were used for sequencing PCR products from positive colonies on an ABI3130 capillary sequencing set-up (Applied Biosystems) using BigDye 3.1 chemistry (Applied Biosystems).
Northern blotting
Ten μg total RNA from tumor samples were separated on a formaldehyde denaturating agarose gel in a morpholino–propane sulphonate (MOPS) buffer and capillary transferred onto a ZetaProbe membrane (Bio-Rad) in 10× SSC for ∼12 h. An exon 3–exon 4 Jdp2 probe generated by PCR using primers Exon3-47 (5′-ctagacgaggaagaagagcgaag-3′) and Exon4-50 (5′-tactacctggtttggcacctctctg-3′) was randomly labeled with [α-32P]dATP and used for hybridization at 65°C in a 0.25 M sodium phosphate buffer/7% SDS, and washed in 0.1× SSC/0.1% SDS. Membranes were finally developed after being exposed to Konica X-Ray AX medical films (Santax Medico) for an appropriate time.
Protein extraction and western blotting
Total protein was extracted on ice from ∼20 mg tissue in a radioimmunoprecipitation assay buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) complemented with 1 mM PMSF and complete Mini, EDTA-free (Roche) protease inhibitor cocktail using a glass homogenizer. Samples were briefly subjected to sonication, incubated on ice for 30 min and protein collected after centrifugation for 30 min at 13 000 × g. Total protein concentrations were determined according to a bovine serum albumin standard curve using the BCA kit (Pierce). Forty (tumor samples) or 25 (tissue samples) μg protein were separated by 15% SDS–PAGE, and electro-transferred onto a 0.2 μM PSQ polyvinylidene fluoride (PVDF)-membrane (Millipore). Membranes were blocked at room temperature in TBS (20 mM Tris–HCl, 200 mM NaCl pH 7.6) containing 5% (w/v) fat-free milk for 1 h, incubated with a primary antibody overnight at 4°C in TBS-T (TBS with 0.05% Tween-20) containing 5% (w/v) fat-free milk (polyclonal anti-Jdp2, 1:1200 dilution, and anti-β-Actin (I-19), 1:6666 dilution Santa Cruz Biotechnology, sc-1616) or TBS-T (monoclonal anti-Jdp2, 1:500 dilution, and monoclonal anti-H2B, 1:5000 dilution), washed three times in TBS-T, incubated with a secondary antibody for 45 min at room temperature in TBS-T with 5% fat-free milk (goat anti-rabbit IgG-HRP, 1:10 000 dilution, Santa Cruz Biotechnology sc-2004; rabbit anti-goat IgG-HRP, 1:5000 dilution, Santa Cruz Biotechnology sc-2768; rabbit anti-mouse IgG-HRP, 1:2000 dilution, DAKO P0260) and finally washed three times in TBS-T. Membranes were developed using ECL PLUS Western Blotting Detection System (GE Healthcare) and exposed to medical film (Konica X-Ray AX medical films Santax Medico). After Jdp2 detection, the membranes were stripped in 25 mM glycine–HCl pH 2, 1% SDS for 15 min and washed several times in TBS before being immunoblotted with an anti-β-Actin antibody. Rabbit polyclonal anti-Jdp2 serum was kindly provided by Dr A. Aronheim, Haifa, Israel (1). The monoclonal antibodies were isolated from hybridoma cell lines J#176-3.2 and J#249-1.1, kindly provided by Dr K. Yokoyama, Ibaraki, Japan (6), using the Nab spin kit (Thermo Scientific).
Vectors
For transient transfections, PCR fragments of Jdp2 full length and isoforms ORFs were cloned into a pSG5-modified pSG5-FLAGnt vector using restriction enzymes XhoI and BamHI. Cloning fragments were generated using forward primers wt-66 (5′-ggggctcgagatgatgcctgggcagatc-3′), 132-64 (5′-ggggctcgagatgtgggtgagaaagggggat-3′), 119-65 (5′-ggggctcgagatgtgtcaggtgggaagctgt-3′) or 107-114 (5′-ggggctcgagatggagtggtgtgtgttga-3′) and reverse primer jdp2stop-67 (5′-ccccggatcctcacttcttgtccagctgctcca-3′). For transduction experiments, PCR fragments were made using appropriate Jdp2 pSG5-FLAGnt vectors as template with forward primer p231-143, 5′-atcgtatgcgtctcccatggactacaaagacgat-3′ and p231-jdp2-145, 5′-atcgtatggacattgctcacttcttgtccag-3′ and inserted into the bicistronic vector AKV-neo-IRES (29). PCR fragments for human NRAS and human NRAS G12D were made using primers p231-144, 5′-atcgtatgccatggactacaaagacgatgacgataaaatgactgagtacaaactggtg-3′, and p231-146, 5′-atcgtatggacattgcttacatcaccacacatg-3′, on appropriate vectors carrying human NRAS or NRAS G12D (kindly provided by Dr. Borja Ballarín-González) and inserted into AKV-neo-IRES. An AvrII-fragment deletion of AKV-neo-IRES lacking the IRES element was used as mock control.
Cell cultures and experiments
NIH 3T3 cells were grown in Dulbecco's Modified Eagle's medium containing glutamax (Gibco) and supplemented with 10% new born calf serum (Invitrogen) and 1% penincillin–streptomycin. Localization studies were conducted by growing NIH 3T3 cells on gelatin-coated glass cover slips and transfecting them using Lipofectamine 2000 (Invitrogen). Cells were then fixed in 4% para-formaldehyde/PBS at room temperature for 10 min, washed in PBS and permeabilized in 0.2% Triton X-100/PBS at room temperature for 10 min. After two washes, cells were left in a blocking solution (0.1% Tween-20/3% BSA/PBS) for 1 h at room temperature. Then, cells were incubated with an anti-FLAG monoclonal antibody (1B11, 1:400 dilution) in the blocking solution for 1 h at room temperature and washed three times in 0.1% Tween-20/PBS. Finally, cells were incubated sheltered from light with a TRITC-conjugated secondary antibody (1:500 dilution) in the blocking solution for 1 h at room temperature before being washed three times in PBS containing 0.1% Tween-20, and two times in PBS. Cover slips were mounted on object glasses using the ProLong antifade mounting solution with DAPI (Invitrogen), and kept at 4°C in dark until analyzed on a Zeiss Axiovert M200 fluorescence microscopy within 24 h.
For cell-growth inhibition analysis, NIH 3T3 cells were transfected as described above and grown for 24 h post-transfection before culturing in puromycin (2.0 μg/ml) for 3 weeks.
Soft agar assay was done by transducing NIH 3T3 cells at a multiplicity of infection (m.o i.) of 1 with AKV-neo-IRES expressing the indicated protein 36 h before seeding 4.2 × 104 into 10 cm2 dishes. Cells were seeded onto a 0.6% bottom layer [complete medium including 0.6% LMP agarose (NuSieve)] and grown in 0.5% top layer for 2 weeks. From each dish, 100 pictures were taken by a Leica DFC 300 FX microscopy and the number of colonies larger than 200 μm were counted using in-house developed software (30).
RESULTS
Integrations into Jdp2
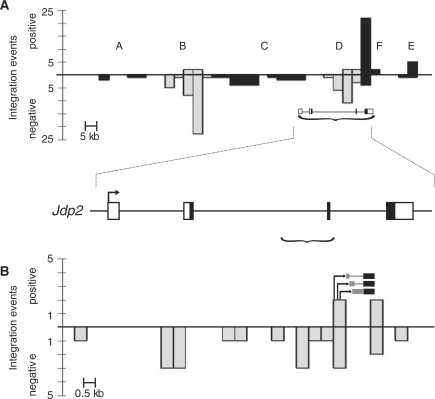
Genomic DNA from tumors induced by SL3-3 murine leukemia virus in 1767 newborn BALB/c mice was isolated and the host-provirus DNA PCR amplified and sequenced. Results from this cohort have previously been published (26,31–33). We wanted to look for integration loci in which the distributions of insertions could indicate novel transcriptional structures and focused on the Jdp2 locus, which is targeted by retroviral insertions in 125 (7%) individual animals (Figure 1A). The locus has previously been described as being an insertional target in murine T-cell lymphomas induced by Moloney MLV (17,19,20) and SL3-3 MLV (18). Retroviral integrations within this locus appeared in clusters, here abbreviated clusters A through F, of which B, D and F were predominant in terms of density and frequency of integrations. Cluster A, B and C were positioned upstream of Jdp2 predominantly in the opposite transcriptional orientation whereas cluster E was positioned downstream of Jdp2. Cluster F consisted of positively orientated proviruses in or near the 3′ UTR in exon 4. In this study, our focus was drawn in particular to cluster D consisting of 25 integrations in the 3′ end of intron 2 of the Jdp2 gene. The predominant inverse transcriptional orientation of the proviruses as compared to Jdp2 suggested a retroviral enhancer activation of a cellular promoter situated in the 3′ end of the integration cluster. The notion was further strengthened by the positive orientation of a few integrations in the 3′ end of cluster D, which could be indicative of promoter activation. To test for such a promoter insertion mechanism of Jdp2 deregulation in tumors carrying a positively orientated provirus within cluster D, we did RT-PCR with primers in the viral U3 region and Jdp2 exon 3 and 4 on tumor cDNA. We indeed found chimeric transcripts consisting of viral and Jdp2 sequences in four out of twenty tumors including one already identified by the initial retroviral tagging (tumor 1363) (Figure 1B). A similar result although at a much higher frequency was found recently by Stewart et al. (20).
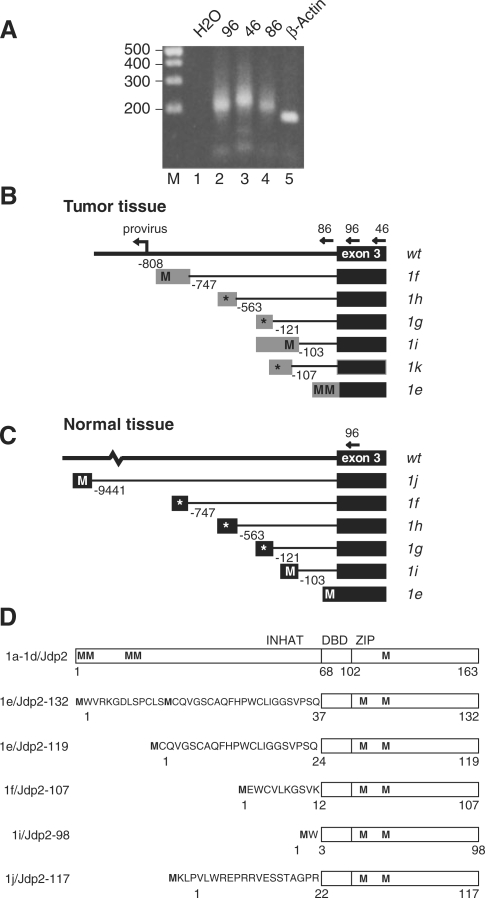
Figure 1.
The Jdp2 locus. (A) The number of integration events around Jdp2 as identified by retroviral tagging are shown in 5000 bp bins from positions 86 370 000 to 86 555 000 on chromosome 12 (UCSC mm8 February 2006) (top). The bars below and above the horizontal axis represent negative and positive, respectively, transcriptional orientation of the proviruses relative to that of the Jdp2 gene. Proviral integration clusters A through F are shown with clusters B and D highlighted in gray. A structure of the Jdp2 gene based on RefSeq NM_030887 is depicted at its chromosomal location (marked with a bracket) with the coding sequence indicated by black coloring (bottom). (B) Close-up on integration cluster D [genome coordinates 86 509 000–86 523 000, marked with a bracket in (A)] in the 3′ end of intron 2 and exon 3 with provirus position and transcriptional orientation depicted as in (A) using 500 bp bins. Schematic examples of provirus-host chimeric transcripts identified are shown with black and gray boxes indicating Jdp2 exon 3 and intron 2 sequences, respectively.
To estimate the frequency of cluster D insertions, we performed directed PCR on control tumors in which no Jdp2 locus integrations were found by retroviral tagging using virus LTR-specific primers and an exon 3 reverse primer. From this we estimated the frequency of integrations into cluster D to be as many as 25% implying subclonality with regard to cluster D insertions, a notion supported by the fact that some control tumors show elevated expression of Jdp2 (see below). Again we found predominant integrations in the opposite orientation as compared to Jdp2. No cluster D insertions in either direction were found in tumors from BALB/c mice infected with the B-cell lymphomagenic Akv MLV signifying the relevance of retroviral Jdp2 intron 2 insertions specifically in T-cell lymphomagenesis (data not shown).
Intragenic integration into Jdp2 correlates with the high expression level of mRNA species including Jdp2 intron 2 sequences
We tested for expressional deregulation of Jdp2 in tumors with MLV insertions within the Jdp2 locus by northern blotting using a probe covering exon 3 and part of exon 4 (Figure 2A). The northern blot analysis included tumors with integrations into cluster D and cluster B (upstream of Jdp2 in opposite orientation), as well as some randomly selected control tumors in which no integration in the Jdp2 locus was detected by the retroviral tagging protocol. In both cluster B and cluster D tumors, the mRNA level of the neighboring genes cFos and Batf were less than three times higher than that observed in control tumors as assessed by quantitative RT-PCR (QRT-PCR) (Figure S1 and data not shown), and hence we conclude that these genes are not targeted by insertions. This is in agreement with the conclusions reached previously by our group (18) and by Hwang et al. (17) using SL3-3 MLV and Moloney MLV, respectively. Consistent with previous results, upregulation of Jdp2 mRNA was observed as compared to control tumors in most cases. However, we note that some tumors with proviral integration into the Jdp2 locus have no Jdp2 upregulation, which could be attributed to tumor sample heterogeneity with many subclonal Jdp2 integrations as mentioned above. The size of the detected Jdp2 mRNA was consistent with the published species (1.5–1.7 kb, see Table 1) (Figure 2B and C). Strikingly, we observed in several cluster D tumors upregulation of small Jdp2 sub-species of which an ∼1.3 kb species was the most common (Figure 2B). These subspecies were not observed in the tumors with integration into cluster B (Figure 2C). We also note expression of larger mRNA species in some tumors.
Table 1.
Features of murine Jdp2 exons 1
| Exona | Coordinateb | Size (bp) | Transcription initiationc | Splice donor sequencec |
|---|---|---|---|---|
| 1a | 86488519 | 199 | gccacagcctGCGGGAGGGA | CTTCTGCACGgtgggttcac |
| 1b | 86488208 | 123 | gggagccgcgGGCGGGGCGC | GGAGGTGAAGgtcagtgggt |
| 1c | 86489121 | 65 | aaccaacaggGCTTCCTTCG | ATATAAGAAGgtaagcaggc |
| 1d | 86491329 | 85 | ctctgtagaaGACATCTTTC | AGCCAAAGAGgtaaaagata |
| 1e | 86520704 | 149 | agctgtgctcAGTTCCACCC | TCTGCAGAGGgtgagtgggg |
| 1e-2 | 86520617 | 236 | tctccgactcAGCAGTCACA | TCTGCAGAGGgtgagtgggg |
| 1f | 86519979 | 23 | atggagtggtGTGTGTTGAA | ATCTGTCAAGgtaagcagga |
| 1f-2 | 86519976 | 26 | agaatggagtGGTGTGTGTT | ATCTGTCAAGgtaagcagga |
| 1g | 86520512 | 116 | agccccagatATTCTGGCTG | GCAGTCACAGgtgtttgctg |
| 1h | 86520110 | 76 | gtcttgctttATGAGTCCTG | GTCCATCCAGgtgagaaggc |
| 1i | 86520625 | 22 | tcagcagtcACAGGTGTTTG | TGAGATGTGGgtgagaaagg |
| 1j | 86511034 | 274 | tcctggttctGCAGACAGCA | AGGGCCAAGGgtgagttcag |
| 1k | 86520513 | 128 | gccccagataTTCTGGCTGA | TTGCTGAGATgtgggtgaga |
aData on exons 1a (RefSeq NM_030887), 1b (mRNA bc019780), 1c (mRNA ab034697) and 1d (EST by317195) are based on UCSC mm 8 February 2006. Minor secondary TSSs found for exons 1e and 1f are denoted 1e-2 and 1f-2, respectively. Exon 1k was only identified in tumor tissue.
bPosition on chromosome 12 of the transcription start site according to UCSC mm8 February 2006.
cExon sequence is in capital case.
To investigate whether the observed 1.3 kb Jdp2 mRNA isoforms in cluster D tumors were generated by transcription from intron 2 as suggested by the integration distribution, we measured the level of canonical versus intronic Jdp2 mRNA (amplicon E2-E3 and I2-E4, respectively, in Figure 2A) with quantitative real-time PCR (QRT-PCR) on a panel of cluster B and cluster D. Tumors without known integrations in the Jdp2 locus as found by retroviral tagging were included as controls (Figure 2D and E). Six out of nine cluster B tumors had >10 times the amount of exon 2–exon 3 Jdp2 mRNA found in control tumors (Figure 2D, bottom). In general accordance with the northern blot, increased mRNA levels were not observed in tumors 959, 150 and 1534. In half of the cluster D tumors (tumors 474, 978, 1908 and 1161), a 5- to 8-fold increase of exon 2–exon 3 mRNA was observed as compared to the controls. In the remaining tumors, mRNA levels were comparable to control tumors (Figure 2D, top). With regard to intronic transcription, an opposite pattern was observed (Figure 2E). Only one cluster B tumor (tumor 645) appeared to have an increased amount of intron 2–exon 4 mRNA as compared to control tumors (Figure 2E, bottom), while five cluster D tumors (tumors 474, 1571, 1833, 1908 and 1161) had 4–45 times higher levels of intronic mRNA (Figure 2E, top). In tumor 978, the high levels of Jdp2 sub-species observed by northern blotting (Figure 2B) was not reflected by amplicon I2-E4 amplification (Figure 2E, top) suggesting a more complex transcriptional structure of Jdp2. Taken together, these data indicate increased transcription of sequence in the 3′ end of intron 2 in tumors harboring intron 2 retroviral integrations (cluster D integrations) compared to tumors with cluster B integrations and tumors with integration outside the Jdp2 locus.
A complex transcriptional structure resides within intron 2 of Jdp2
Two immediate explanations can account for the expression of mRNA including the 3′ end of Jdp2 intron 2. One possibility is that transcription starts upstream of exon 2 and utilizes a cryptic splice acceptor site in the 3′ end of intron 2. An alternative hypothesis would invoke the existence of alternative promoters within intron 2.
We tested for alternative splice donor sites with PCR on cDNA from a range of tumors using forward primers situated in upstream Jdp2 exons together with reverse primers in the 3′ end of intron 2. However, we repeatedly failed to find cDNA representing alternative splicing between any of the four possible exons 1 (supported by transcription data on http://genome.ucsc.edu/) and the 3′ end of intron 2 (data not shown).
To search for a possible novel transcription start site immediately upstream of exon 3, we performed 5′ RACE on RNA from tumor tissue. We used the tumor sample identified as having the highest level of intron 2 containing mRNA among the tumors as determined by QRT-PCR (tumor 1161, 45-fold higher as compared to control tumors, see Figure 2E). In addition, we performed 5′ RACE on RNA from bone marrow (BM) and whole brain tissue from uninfected BALB/c mice. During SL3-3 MLV infection of newborn mice, BM cells are a likely reservoir for early integration events whereas brain tissue is unrelated to infection. 5′ RACE PCR on tumor tissue was done with either of three reverse primers (oligo 86 locates in intron 2, and oligo 96 and 46 are specific to exon 3), whereas 5′ RACE on BM and brain tissue was done using a single primer (oligo 96) (see Figure 3). As a control, 5′ RACE PCR was done with a reverse primer in the 5′ end of the gene for β-Actin (Actb). PCR products were cloned and between 50 and 100 bacterial colonies from each 5′RACE PCR representing individual transcription start sites (TSS) sequenced. The result of the analysis is summarized in Table 1. As expected, 5′ RACE PCR for Actb yielded a sharp band in agreement with a single predominant transcription initiation site (Figure 3A, lane 5). In contrast, we always detected smeared bands in both the Jdp2 intron 2 and exon 3-specific 5′RACE PCRs suggesting existence of multiple 5′ ends for the intron 2 including mRNA species (Figure 3A, lanes 2–4).
Figure 3.
Identification of Jdp2 intron 2 mRNAs. (A) Ethidium bromide-stained agarose gel showing representative PCR products with linker-specific forward primer on 5′ RACE cDNA from tumor 1161 using different reverse primers in Jdp2 (oligos 96, 46 and 86, lanes 2, 3 and 4, respectively) and Actb exon 3 (lane 5); in lane 1 no cDNA template was added. (B) and (C) Schematic structure of the alternative Jdp2 exon 1e through 1k as found by 5′ RACE in tumor tissue (B) and normal tissue (C). Positions of exon 1-specific splice donor sites relative to exon 3 are given in base pairs. Putative start codons (M) in frame with the ORF of Jdp2 are indicated, while an asterisk indicates that no ORF is present in frame with Jdp2. (D) Protein structure of Jdp2 as generated from exon 1a through 1d, and predicted Jdp2 isoforms generated from exon 1e, 1f, 1i and 1j. The INHAT domain as well as the basic DNA binding domain (DBD) and the leucine zipper (ZIP) regions are indicated. Methionines are indicated (M) and the N-terminal peptides are shown for the isoforms.
When using an intron 2-specific 5′ RACE PCR on tumor tissue we found several TSSs within a 110 nucleotide window located ∼150 base pair upstream of exon 3. Transcripts from this region all continue without splicing into exon 3 giving rise to mRNA species with an alternative intron 2-derived 5′ terminus in place of the normal exon 1 and exon 2 of Jdp2 (corresponding to exon 1e in Figure 3B).
TSSs cloned from the exon 3-specific 5′ RACE PCR identified the same 110 window as found above. In addition, a variety of transcripts utilizing intron 2 splice donor sites and the canonical exon 3 splice acceptor site were identified. The transcripts were denoted exons 1e, 1f, 1g, 1h, 1i, 1j and 1k based on their splicing structure (Table 1 and Figure 3B and 3C). Two less abundant initiation sites upstream of the major initiation sites in exon 1e (brain tissue) and 1f (bone marrow) were denoted 1e-2 and 1f-2, respectively. We reserve the terms exons 1a through 1d for the four alternative exons 1 that currently are supported by public transcript records (http://genome.ucsc.edu/), and which are all situated outside the region considered here (see Table 1). While exon 1k was exclusively observed in tumor tissue, exon 1j, situated in intron 2 almost 10 kb upstream of exon 3, was found only in normal tissue (brain). The far most predominant transcripts in all three tissues were exons 1f and 1e accounting for 74% (tumor tissue), 88% (brain) and 98% (bone marrow) of all transcripts. Since the translation initiation codon of Jdp2 is encoded by exon 2, the alternative subspecies identified here could produce protein isoforms with an altered N-terminus. Interestingly, a methionine codon in frame with Jdp2 was present in exons 1e, 1f and 1i from tumors as well as in exon 1j from brain (Figure 3B and C). Transcription from exon 1e may give rise to either a 132-amino-acid isoform, Jdp2-132, or, using a second methionine codon, a 119-amino-acid isoform, Jdp2-119. Transcription from exons 1f, 1i and 1j generates Jdp2 isoforms of 107-, 98- and 117-amino-acid, respectively. In this line, it should be noted that exon 2 of Jdp2 encodes a protein domain responsible for the inhibition of histone acetylation (7). Except for one sequence (a brain exon 1e sequence), exon 1e and exon 1f TSS tags from normal tissue are situated 3′ to the methionine codons and hence cannot give rise to Jdp2-132/119 or Jdp2-107 production. Transcription in tumor tissue, in contrast, initiated close to the integrated provirus and mostly upstream of the positions found in normal tissue. Thereby, the provirus-activated transcripts disclose upstream Jdp2-in-frame methionine codons (compare exons 1e and 1f in Figure 3B with Figure 3C). Figure 3D summarizes the different predicted protein structures.
In support of our data, we note that all eight different TSS positions identified in uninfected tissue had a pyrimidine-purine dinucleotide at the −1/+1 transcription start site positions (CpA, TpA or TpG in Table 1), in line with the accepted view on the initiator motif (Inr) in mammalian polymerase II transcribed genes (34,35). In tumor tissue, 32% of the cloned TSSs were non-PyPu dinucleotides as exemplified in exon 1k (Table 1 and data not shown).
We conducted a bioinformatic analysis to look for cis-regulatory elements in 1200 base pairs covering the region between 300 bp 5′ to the TSS of exon 1f and exon 3 using the DiAlign software (36) (Figure S2). The region is not conserved between mammals and avian species since the similarity between mouse and chicken was confined to the exon 3 sequence (data not shown). As would be expected, the relative conservation score is generally higher when only including mouse and rat; larger patches (50–150 bp) of differences exist in chimpanzee (insertion), dog (insertion) and cow (insertion and deletion) and in human, chimpanzee, macaque, cow and dog as a group (insertion) as compared to mouse and rat (data not shown). Despite this, minor peaks of conservation when considering all species are found around the transcriptional initiation regions of exons 1e, 1f and 1g (Figure S2). When only considering mouse and rat, conservation peaks are seen on all the transcription start sites of the alternative exon 1s in the region (Figure S2). Other potential start sites of transcription or regulatory modules such as enhancers or splice regulatory sequences may reside within peaks of conservation, which are not overlapping the TSSs found here. To this end, we used MatInspector to identify possible conserved transcription factor-binding motifs within the considered regions (37) (Figure S3–S5). The regions indeed have several conserved transcription factor motifs including binding sites for SP1, YY1 and ETS factors. We note the absence of obvious conserved TATA-box sequences within the sequence immediately in front of the TSS sequences.
Jdp2 protein isoforms are expressed in T-cell tumor tissue and normal tissue
The identification of multiple mRNA forms with the potential to encode for novel protein isoforms of Jdp2 prompted us to investigate mRNA levels of the abundantly found alternative transcripts 1e-3-4 and 1f-3-4 in comparison with Jdp2 protein levels. QRT-PCR specific for exon 1e and specific for the splicing between exons 1f and exon 3 was done with amplicons E1e-E4 and E1f-E4, respectively (Figure 2A). Amplicon E1e-E4 lies downstream of the two AUG codons and thus does not distinguish the longer and shorter 1e exons (exons 1e-2 and 1e in Table 1). Figure 4A–C shows the results from some of the most differentially expressed tumor samples as compared to the normal thymus expression (set to 1). Of the three cluster D tumors (763, 1363 and 1161), tumor 763 had >100-fold increased levels of the canonical wild-type transcript similar to the cluster B tumors (20 and 181) (Figure 4A). As suggested by northern blotting (Figure 2C), control tumor 1046 also displayed highly increased wild-type levels. PCR using virus and cluster D-specific primers in tumor 1046 in fact detected integration into cluster D while the proviral status of cluster B was not investigated. Levels of the 1e-3-4 RNA were >50 times higher in the three cluster D tumors and tumor 1046, whereas cluster B tumors only displayed a moderate increase (∼10-fold) as compared to control thymus (Figure 4B). Expression of the 1f-3-4 RNA was >1000-fold higher in tumors 1363 and 1161 while between 50 and 150 times higher in tumors 763, 20, 181 and 1046 as compared to control thymus.
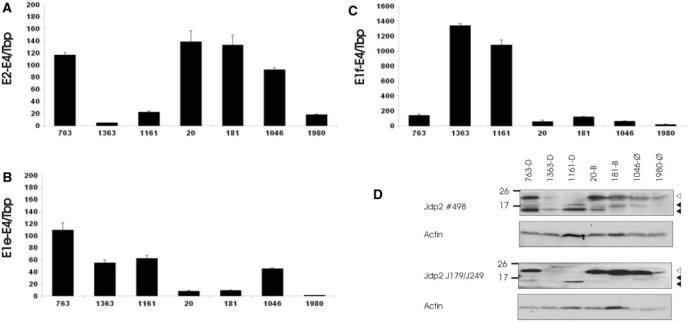
Figure 4.
Jdp2 isoforms are highly expressed in T-cell tumors. (A–C) QRT-PCR was done in triplicates on tumor tissues to detect full length (A), exon 1e-3-4 (B) and exon 1f-3-4 (C) mRNA. Expression signal was normalize with Tbp and shown as fold difference to uninfected control thymus. The figures are representative of two to three experiments. (D) Two separate western blot experiments using 0.2 μM PVDF membranes were done on crude protein extracts from the same tumors as in (A–C) using either polyclonal anti-Jdp2 antibody (#489) (top) or a mix of monoclonal anti-Jdp2 antibodies J176 and J249 (bottom). Membranes were subsequently immunoblotted using anti-β-Actin antibody. Open and closed arrows indicate position of full length (∼20 kDa) and isoform Jdp2 (∼14 and 17 kDa), respectively.
Jdp2 protein levels in the same tumor samples were then investigated by western blotting using a rabbit polyclonal anti-Jdp2 protein (Figure 4D, top). The relative signal of full length Jdp2 (∼20 kDa) between tumors corresponded to the relative exon 2-3 mRNA levels. Smaller bands (at ∼17 and 14 kDa) were detected in all samples except control tumor 1980 with the highest signal in tumors 763 and 1161. To confirm that the smaller bands represent alternative Jdp2 isoforms, the same protein samples were subjected to a separate SDS–PAGE immunoblot using Jdp2 murine monoclonal antibodies (Figure 4D, bottom). Full-length Jdp2 was readily detected in the same samples as when using the polyclonal antibody. Further, in samples 763 and 1161, a band corresponding to the lower 14 kDa Jdp2 isoform was evident as was, although more weakly, the 14 kDa band in sample 20 and the 17 kDa band in samples 763, 1161 and 1046. Tumor 1363 is a special case in which the provirus is transcriptionally orientated similar to Jdp2. This allows for the formation of chimeric spliced or unspliced mRNAs containing viral sequences, which may have an impact on the ratio between levels of protein isoforms and RNA (compare Figure 4B and C with D for tumor 1363). Together with the mRNA expression data, these data support the notion of the expression of Jdp2 isoforms in T-cell tumor tissue.
We next analyzed the expression level of canonical Jdp2 and truncated Jdp2 in a panel of tissues from BALB/c mice not infected with MLV. In order to minimize effects from sample heterogeneity, relative expression signal was normalized to the geometric mean of Actb and Tbp (28) (Figure 5A–C, black bars). In some cases, this had a dramatic effect on the relative expression level as compared to normalization to total RNA (38,39) (Figure 5B, gray bars), as is evident, for instance, for amplicon E1e–E4 in testis and brain. The following is based on interpretation using Tbp and Actb normalizations. Full-length Jdp2 mRNA was relatively highly expressed in the brain and lung tissue, while an intermediate expression level was seen in spleen, heart, bone marrow and kidney (Figure 5A). The expression of exon 1e was relatively highest in the brain (Figure 5B), while exon 1f was most prominently expressed in heart tissue (Figure 5C). We also estimated the absolute copy number of transcripts by using standard curve generated from Jdp2 amplicon-containing plasmids of known concentrations (see ‘Materials and Methods’ section). Canonical Jdp2 E2-E4 was 6 (in testis) to 100 (in heart) times more abundant than Jdp2 E1e-E4, and between 3 (in liver) and 400 (in lung) times more abundant than Jdp2 E1f-E4 (data not shown). Next, we did western blot analysis using a polyclonal anti-Jdp2 antibody on tissue samples extracted from BALB/c mice (Figure 5D). Immunoblotting against β-Actin and histone H2B (a marker for nuclear content) was subsequently done to evaluate for protein loading. Highest full-length Jdp2 signal was seen in the skeletal muscle and, to a lesser extent, in testis, heart and liver. A smaller band of similar size as the larger one seen in tumor samples (∼17 kDa) was most prominently observed in heart although also in testis, liver and skeletal muscle. In liver tissue, a protein of smaller size was evident. Using monoclonal antibodies, however, we could only detect the full-length Jdp2 in the skeletal muscle upon long exposure time (data not shown). This may be due to the relatively low expression level as compared to tumor samples with integrations in combination with lower reactivity of monoclonal antibodies. In summary, these data indicate that Jdp2 intragenic MLV insertions upregulate Jdp2 isoforms from intron 2 promoters, which are active in various non-infected murine tissues.
Jdp2 isoforms localize to the nucleus similar to full-length Jdp2
Jdp2 localizes to the nucleus when expressed from a heterologous promoter specifically in the heart (15), yet any nuclear localization signal remains undefined. The Jdp2 isoforms identified here have the N-terminal part replaced by a short peptide encoded by the various alternative exons 1. To investigate the subcellular localization of Jdp2 and to see if the Jdp2 isoforms had an altered localization pattern, full length as well as Jdp2-132, Jdp2-119 and Jdp2-107 was cloned into a pSG5-derived vector, pFLAGnt, and expressed in mouse fibroblast as N-terminal FLAG fusion proteins. Protein localization was then visualized by immunofluorescence using an anti-FLAG antibody. Full-length Jdp2 localized exclusively to the nucleus (Figure 6A). The same localization was observed for isoforms Jdp2-132, Jdp2-119 and Jdp2-107 (Figure 6B–E) whereas no signal was seen in cell transfected with the same vector but without Jdp2 transgene (Figure 6F). We note that in some Jdp2-132 transfected cells, increased signal intensity was observed asymmetrically near the nuclear rim (Figure 6B). This localization pattern was not a consequence of high expression of FLAG-Jdp2-132 as the proportion of cells with this localization pattern was equal between high and low level Jdp2-132 expressing cells.
Ectopic expression of Jdp2 and Jdp2 isoforms increase tumorigenic potential of oncogenic NRAS G12D
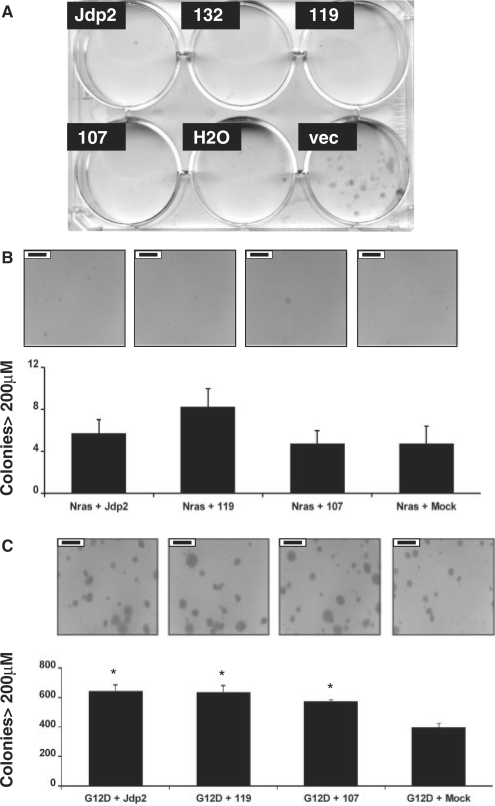
The role of Jdp2 in apoptosis and cell cycle control is subject to debate and seems to vary with cellular context (10,12,13,21). To test if the cell growth inhibitory effect ascribed previously to full-length Jdp2 also was present for the 132-, 119- and 107-amino-acid isoforms we transfected NIH3T3 cells with the respective expression vectors and added puromycin 24 h post-transfection to select for the capacity to form colonies. Whereas we readily observed colonies among cells transfected with the empty plasmid (pFLAG), cells transfected with Jdp2, Jdp2-132, Jdp2-119 or Jdp2-107 died (Figure 7A). Importantly, the ectopic expression level was several orders of magnitude higher than that observed in the tumor samples (Figure S6). Thus, Jdp2-mediated cell growth inhibition under these conditions is not only restricted to the full-length Jdp2 isoform but is also evident for protein isoforms lacking the N-terminal INHAT domain.
Figure 7.
Phenotypic effects of ectopic expression of Jdp2 isoforms. (A) NIH3T3 cells were transiently transfected with vectors expressing N-terminally FLAG tagged full length Jdp2 or either of three N-terminally Jdp2 isoforms, Jdp2-132, Jdp2-119 or Jdp2-107. Twenty-four hours after transfection, cells started growing in a selective medium containing puromycin. Three weeks later, the cells were fixed in methanol and colonies stained with methylene blue. The results are representative of two independent experiments, each done in duplicate. (B and C) NIH 3T3 cells were co-transduced with the indicated vectors (m.o.i. = 1) 48 h before being seeded in the medium supplemented with 0.5% agar. After 2 weeks of growth, 100 pictures per dish were taken, and the number of colonies larger than 200 μm were counted. Error bars indicate the standard error of the mean from four independent experiments of which representative pictures are shown. The black scale bar—insert in each pictures—corresponds to 500 μm. A significant difference (P ≤ 0.05) to ‘G12D + Vector’ by two-tailed Student's t-test is shown with an asterisk.
Since full-length Jdp2 has been reported to inhibit RasV12-induced focus formation (21), we tested the effect Jdp2 and Jdp2 isoforms on anchorage-independent growth in the context of human NRAS and oncogenic NRAS G12D. In order to minimize possible apoptotic effects observed in Figure 7A, Jdp2, Jdp2-119 and Jdp2-107 was expressed from an internal ribosomal entry site (IRES) within the LTR-driven retroviral vector AKV-Neo-IRES. Figure S7 demonstrates that the transduced cells exhibit moderate over-expression as compared to endogenous full-length Jdp2, while a more prominent over-expression was observed under the transiently expressing conditions used in Figure 7A. NIH 3T3 fibroblasts co-transduced with a retroviral vector expressing NRAS and a retroviral vector expressing Jdp2 or either isoforms did not sustain colony growth significantly different from the NRAS-expressing vector alone when seeded in a complete medium supplemented with 0.5% agar (Figure 7B). Expression of Jdp2, Jdp2-119 or Jdp-107 alone also did not result in colony formation significantly different from either mock transduction or NRAS transduction (data not shown). In contrast, when we co-transduced NRAS G12D together with either Jdp2 forms, a significant increase of ∼50% in colony formation was seen as compared to NRAS G12D alone (P < 0.05, two-tailed Student's t-test) (Figure 7C). Thus, both Jdp2 proteins with and without the N-terminal part harboring the INHAT activity can increase transformation by oncogenic NRAS G12D but not NRAS.
DISCUSSION
Here, we have described the identification of a novel region within the second intron of the AP-1 repressor Jdp2 from which several alternative mRNA species are expressed in T-cell lymphomas of mice and in normal tissue. Some, but not all, of the alternative mRNAs encode for protein N-terminal isoforms of Jdp2.
The existence of intragenic promoter activity could be hypothesized from the distribution of retroviral integrations in the Jdp2 locus as found by retroviral tagging on T-cell tumors induced by SL3-3 MLV in 1767 mice. When integrated in opposite transcriptional orientation of the proto-oncogene being activated the provirus is typically situated upstream of the cellular promoter. This mechanism has been termed ‘enhancer activation’ and is exemplified by activation of Gfi1 (17,40), Rras2 (41,42) and Rasgrp1 (20,24) in various models (data from (43)), as well as integration clusters A, B and C reported here and elsewhere (17,18). By analogy, oppositely orientated proviruses within the transcriptional unit of a cellular gene such as cluster D in Jdp2 may reveal novel promoters 3′ to the integrations. A recent publication from our group on insertional activation of Bach2 in B-cell lymphomas (44), as well as unpublished data, indicates a more general phenomenon of proviral activation of ‘hidden’ intragenic promoters, and we propose that large-scale transcription start site mapping in provirally targeted genes will contribute in finding new structures of already known cancer genes. Using existing retroviral tagging data such as from RTCGD (43) additional promoter regions are envisioned to be identified. It would be of interest to investigate whether such alternative transcripts have a regulated expression in normal tissue, as in case of Jdp2, or require cis-regulatory elements such as from provirus insertion or chromosomal translocation.
It is now evident that the transcriptome of higher mammals is far more complex than earlier anticipated (45–50). A significant contribution to increased transcriptional complexity comes from the usage of alternative promoters and accordingly first exons among polymerase II transcribed genes (34,51–53). Current database records (http://genome.ucsc.edu/) support the existence of four non-overlapping initiating non-coding exons of Jdp2 that splice to a common splice acceptor site in exon 2 (exons 1a–1d in Table 1), although it should be kept in mind that the precise transcription start sites of exons 1a, 1b and 1d await final verification. The novel Jdp2 transcript structures identified here within intron 2 add to the complexity of Jdp2 transcriptional regulation. All lack TATA-box consensus elements surrounding the 30 bp upstream regions from the TSS. This result is not unexpected according to recent transcriptome analyses showing that the presence of TATA-box sequences is an exception rather than a rule and that numerous alternative initiation sequences must exist (34,54–56).
The transcripts including Jdp2 intron 2 sequences constituted either a 5′ extension of a common exon (in case of exon 1e) or mutually exclusive exons inside an existing intron (in case of exons 1f through 1k) and belong to the most common classes of alternative first exons among both humans and mice (51,57). We have by RT-PCR on mouse tumor tissue identified potential intron 2-initiating exons in addition to those reported here by 5′ RACE, as well as one in the region harboring the known exons 1a through 1d (data not shown). We do not at present know if these are expressed in normal tissue as well. Generally, alternative promoter usage and subsequent use of alternative exons 1 is related to tissue- and temporal-specific expressions (58–63). The molecular regulation of Jdp2 transcription is largely unknown except for the observation that several uncharacterized transcripts of various sizes have been described in different tissues (2,18). It is thus tempting to connect the identified complex transcription pattern of Jdp2 with the regulated expression of full-length and truncated transcripts. Although we have not addressed Jdp2 regulation specifically, the presented expression analyses indicate tissue-specific expression of both canonical and alternative Jdp2 mRNAs. For instance, while skeletal muscle expresses relatively high levels of both canonical and alternative transcripts 1e-3-4 and 1f-3-4 Jdp2, in brain tissue levels of 1f-3-4, were relatively low while canonical and 1e-3-4 Jdp2 levels were high. Along this line, the absolute copy number of isoform transcripts was judged to be in the similar range as the canonical mRNA for some tissues, such as Jdp2 1f-3-4 in skeletal muscle, which may indicate a specific role in this tissue. Interestingly, Jdp2 has been implicated in the differentiation of C2 myoblast cells (10).
Several factors influence the discrepancies observed between mRNA and protein signal in some tissues such as testis. First, we focused on the two most abundantly expressed alternative transcripts as found in one tumor and two normal tissue samples (bone marrow and brain) by 5′ RACE, and acknowledge that our expression analyses were far from exhaustive. Hence, some of the observed protein species are likely to derive from sequences other than exons 1e and 1f. Our 5′ RACE analysis intriguingly showed most alternative exons 1e and 1f transcripts in bone marrow and brain to be initiated after the start codons that were disclosed in tumor tissue. Although an in-frame methionine codon exists in exon 4 allowing for production of a 54-amino-acid peptide by these transcripts, any biological function of such peptide is not obvious since it would lack part of the leucine zipper domain of Jdp2. It should be mentioned, however, that ATF3, which is the bZIP transcription factor most related to Jdp2 (64), encodes a naturally occurring splice variant ATF3ΔZip lacking the leucine zipper domain (65). Despite lacking the ability to bind DNA, this isoform antagonized ATF3 repression in a way that was speculated to involve sequestering of inhibitory co-factors (65). Second, different involvement of post-transcriptional regulation in various tissues is expected. Finally, the sequence around the start AUG codon differs among the investigated transcripts. According to the most coherent model for eukaryotic translation initiation ribosome preinitiation components are recruited near the 5′ cap of the mRNA and scan the 5′UTR for the first AUG codon to start translation (reviewed in (66)). According to phylogenetic and genetic evidence (67–71) the ‘optimal’ nucleotide context in terms of translation initiation efficiency is GCC(A/G)CCAUGG (bold nucleotides are envisaged to be the most important). From this the AUG context in exon 1f (UGGAGAAUGG) would be predicted to be considerably stronger than in the two encoded by exon 1e (GCUGAGAUGU for Jdp2-132, and CUGUCCAUGU for Jdp2-119). It will be important to thoroughly characterize the expression pattern of Jdp2 alternative exons 1 under various stress stimuli known to involve Jdp2 function.
Since little is known about the subcellular localization of Jdp2 (15), we compared the localization pattern of full-length and isoform Jdp2 by immunofluorescence. We also considered the possibility that the alternative N-termini of the isoforms, and in particular the long alternative N-terminus of Jdp2-132, could harbor signals involved in subcellular localization. However, both Jdp2 and Jdp2-132, Jdp2-119 and Jdp2-107 isoforms accumulated exclusively to the nucleus on over-expression in murine fibroblasts. Hence, our results indicate that the N-terminus of neither Jdp2 (residues 1-68) nor of the isoforms contain important determinants involved in nuclear transport. These, then, are likely to locate within the basic DNA-binding region as for other AP-1 factors.
Retroviral activation of Jdp2 in T-cell lymphomas of mice is currently the strongest evidence for a gain-of-function potential of Jdp2 in cancer development in the hematopoietic system. The observation that truncated transcripts in some tumors were almost exclusively up-regulated indicate a tumorigenic potential of Jdp2 isoforms lacking the N-terminus, which is involved in making associations to histones and contributes a histone acetylation inhibitory (INHAT) capacity to Jdp2 (7). Other tumor samples (mainly cluster B tumors) apparently resulted in elevation of only canonical Jdp2. These observations are in line with the finding of increased anchorage-independent growth potential of NRAS G12D both when expressed together Jdp2 and Jdp2 isoforms lacking the INHAT. It should be noted that in contrast to our findings, Heinrich et al. (21) reported full-length Jdp2 to repress oncogenic RasV12 transformation (21). We speculate that a possible explanation could be related to highly different ectopic levels of Jdp2; by applying a LTR/IRES-mediated expression strategy after retroviral transduction, we dramatically lowered ectopic Jdp2 protein levels as compared to SV40 promoter-driven expression after plasmid transfection, and hence may have been below a certain threshold for cellular toxicity responses, or minimized squelching of Jdp2-dimerization partners by ectopic Jdp2 not bound to DNA (72). In our clonogenic assays, which were based upon plasmid transfection of SV40 promoter-driven Jdp2 expression constructs followed by selection for high transgene expression, we observed lack of cell survival in the case of full-length as well as isoform Jdp2 proteins. This result independently demonstrates an effect of Jdp2 isoforms lacking the N-terminal INHAT domain.
The finding of a similar potential of full-length and truncated Jdp2 in colony formation in the presence of Nras G12D raises the possibility that the tumorigenic role of retroviral activation of Jdp2 is independent of the N-terminal repressive domain. Since some tumors samples had low canonic RNA levels but high intron 2-derived transcripts (e.g. tumors 1161 and 1363), such a scenario would predict that selection for MLV integrations within Jdp2 be attributed solely to the ability to activate the intron 2 promoters. An alternative possibility is that full-length and truncated Jdp2 have separate roles during T-cell lymhomagenesis. In favor of this model are several indirect connections between transformation and the Jdp2 N-terminus. For instance, Blazek et al. (16) found the N-terminus of Jdp2 indispensable for transformation of chick embryo fibroblasts and proposed the Jdp2-Fos heterodimer as a likely candidate for this activity (16). Also, the N-terminus, together with the DNA-binding domain of Jdp2, was important in Jdp2-stimulated progesterone-dependent transcriptional activation by PR (8), linking this part of Jdp2 to the homeostasis of female reproductive tissue. Recently, Jdp2 was found to repress transcription through consensus ATF/CRE sites of the immediate early gene ATF3 (73), which in some models appears to act as a tumor suppressor of Ras-mediated tumorigenesis (74,75). Although the involvement of Jdp2 N-terminus was not specifically addressed, based on well-established work on Jdp2 transcription repression through ATF/CRE (1,2,7,16), ATF3 repression is likely to require the INHAT domain of Jdp2 also. It thus remains to be clarified whether Jdp2 isoforms lacking an inhibitory N-terminal domain but have retained the basic and zipper domain encoded by exons 3 and 4 contribute a dominant negative or a loss of function effect to cellular Jdp2-including protein complexes. In that regard, it would be important to investigate how a deregulated balance of Jdp2 isoforms, as found here in T-cell lymphomas, impacts on AP-1 complex constituents and Jdp2 target genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Danish Cancer Society, Dansk Kræftforsknings Fond and The Danish Agency for Science, Technology, and Innovation; in part by a scholarship from the Danish Research School in Molecular Cancer Research (to M.H.R.); a Hallas–Moller fellowship from the NovoNordisk Foundation (to A.L.N.); NIH grant R01A141570 (to M.W.). Funding for open access charge: Danish Agency for Science, Technology, and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Ami Aronheim and Dr Kaz Yokoyama for anti-Jdp2 antibodies, and to Dr Borja Ballarín-González for providing Nras vectors. They also thank Lone Højgaard Nielsen for excellent technical assistance.
REFERENCES
- 1.Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol. Cell Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin C, Ugai H, Song J, Murata T, Nili F, Sun K, Horikoshi M, Yokoyama KK. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 2001;489:34–41. doi: 10.1016/s0014-5793(00)02387-5. [DOI] [PubMed] [Google Scholar]
- 3.Broder YC, Katz S, Aronheim A. The ras recruitment system, a novel approach to the study of protein–protein interactions. Curr. Biol. 1998;8:1121–1124. doi: 10.1016/s0960-9822(98)70467-1. [DOI] [PubMed] [Google Scholar]
- 4.Cherasse Y, Chaveroux C, Jousse C, Maurin AC, Carraro V, Parry L, Fafournoux P, Bruhat A. Role of the repressor JDP2 in the amino acid-regulated transcription of CHOP. FEBS Lett. 2008;582:1537–1541. doi: 10.1016/j.febslet.2008.03.050. [DOI] [PubMed] [Google Scholar]
- 5.Weidenfeld-Baranboim K, Bitton-Worms K, Aronheim A. TRE-dependent transcription activation by JDP2-CHOP10 association. Nucleic Acids Res. 2008;36:3608–3619. doi: 10.1093/nar/gkn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin C, Li H, Murata T, Sun K, Horikoshi M, Chiu R, Yokoyama KK. JDP2, a repressor of AP-1, recruits a histone deacetylase 3 complex to inhibit the retinoic acid-induced differentiation of F9 cells. Mol. Cell Biol. 2002;22:4815–4826. doi: 10.1128/MCB.22.13.4815-4826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin C, Kato K, Chimura T, Yamasaki T, Nakade K, Murata T, Li H, Pan J, Zhao M, Sun K, et al. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat. Struct. Mol. Biol. 2006;13:331–338. doi: 10.1038/nsmb1063. [DOI] [PubMed] [Google Scholar]
- 8.Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol. Cell Biol. 2002;22:5451–5466. doi: 10.1128/MCB.22.15.5451-5466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaida R, Ohtsuka T, Okutsu J, Takahashi T, Kadono Y, Oda H, Hikita A, Nakamura K, Tanaka S, Furukawa H. Jun dimerization protein 2 (JDP2), a member of the AP-1 family of transcription factor, mediates osteoclast differentiation induced by RANKL. J. Exp. Med. 2003;197:1029–1035. doi: 10.1084/jem.20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrovsky O, Bengal E, Aronheim A. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J. Biol. Chem. 2002;277:40043–40054. doi: 10.1074/jbc.M205494200. [DOI] [PubMed] [Google Scholar]
- 11.Nakade K, Pan J, Yoshiki A, Ugai H, Kimura M, Liu B, Li H, Obata Y, Iwama M, Itohara S, et al. JDP2 suppresses adipocyte differentiation by regulating histone acetylation. Cell Death Differ. 2007;14:1398–1405. doi: 10.1038/sj.cdd.4402129. [DOI] [PubMed] [Google Scholar]
- 12.Lerdrup M, Holmberg C, Dietrich N, Shaulian E, Herdegen T, Jaattela M, Kallunki T. Depletion of the AP-1 repressor JDP2 induces cell death similar to apoptosis. Biochim. Biophys. Acta. 2005;1745:29–37. doi: 10.1016/j.bbamcr.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Piu F, Aronheim A, Katz S, Karin M. AP-1 repressor protein JDP-2: inhibition of UV-mediated apoptosis through p53 down-regulation. Mol. Cell Biol. 2001;21:3012–3024. doi: 10.1128/MCB.21.9.3012-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakade K, Pan J, Yamasaki T, Murata T, Wasylyk B, Yokoyama KK. JDP2 (Jun dimerization protein 2)-deficient mouse embryonic fibroblasts are resistant to replicative senescence. J. Biol. Chem. 2009;284:10808–10817. doi: 10.1074/jbc.M808333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehat I, Heinrich R, Ben-Izhak O, Miyazaki H, Gutkind JS, Aronheim A. Inhibition of basic leucine zipper transcription is a major mediator of atrial dilatation. Cardiovasc. Res. 2006;70:543–554. doi: 10.1016/j.cardiores.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Blazek E, Wasmer S, Kruse U, Aronheim A, Aoki M, Vogt PK. Partial oncogenic transformation of chicken embryo fibroblasts by Jun dimerization protein 2, a negative regulator of TRE- and CRE-dependent transcription. Oncogene. 2003;22:2151–2159. doi: 10.1038/sj.onc.1206312. [DOI] [PubMed] [Google Scholar]
- 17.Hwang HC, Martins CP, Bronkhorst Y, Randel E, Berns A, Fero M, Clurman BE. Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc. Natl Acad. Sci. USA. 2002;99:11293–11298. doi: 10.1073/pnas.162356099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen MH, Sørensen AB, Morris DW, Dutra JC, Engelhard EK, Wang CL, Schmidt J, Pedersen FS. Tumor model-specific proviral insertional mutagenesis of the Fos/Jdp2/Batf locus. Virology. 2005;337:353–364. doi: 10.1016/j.virol.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 19.Sauvageau M, Miller M, Lemieux S, Lessard J, Hebert J, Sauvageau G. Quantitative expression profiling guided by common retroviral insertion sites reveals novel and cell type-specific cancer genes in leukemia. Blood. 2008;111:790–799. doi: 10.1182/blood-2007-07-098236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart M, Mackay N, Hanlon L, Blyth K, Scobie L, Cameron E, Neil JC. Insertional mutagenesis reveals progression genes and checkpoints in MYC/Runx2 lymphomas. Cancer Res. 2007;67:5126–5133. doi: 10.1158/0008-5472.CAN-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinrich R, Livne E, Ben-Izhak O, Aronheim A. The c-Jun dimerization protein 2 inhibits cell transformation and acts as a tumor suppressor gene. J. Biol. Chem. 2004;279:5708–5715. doi: 10.1074/jbc.M307608200. [DOI] [PubMed] [Google Scholar]
- 22.Katz S, Aronheim A. Differential targeting of the stress mitogen-activated protein kinases to the c-Jun dimerization protein 2. Biochem. J. 2002;368:939–945. doi: 10.1042/BJ20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz S, Heinrich R, Aronheim A. The AP-1 repressor, JDP2, is a bona fide substrate for the c-Jun N-terminal kinase. FEBS Lett. 2001;506:196–200. doi: 10.1016/s0014-5793(01)02907-6. [DOI] [PubMed] [Google Scholar]
- 24.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 25.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 26.Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M, Killeen N, Wabl M. Activation of an oncogenic microRNA cistron by provirus integration. PNAS. 2006;103:18680–18684. doi: 10.1073/pnas.0609030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahrami S, Jespersen T, Pedersen FS, Duch M. Mutational library analysis of selected amino acids in the receptor binding domain of envelope of Akv murine leukemia virus by conditionally replication competent bicistronic vectors. Gene. 2003;315:51–61. doi: 10.1016/s0378-1119(03)00719-4. [DOI] [PubMed] [Google Scholar]
- 30.Lovmand J, Justesen J, Foss M, Lauridsen RH, Lovmand M, Modin C, Besenbacher F, Pedersen FS, Duch M. The use of combinatorial topographical libraries for the screening of enhanced osteogenic expression and mineralization. Biomaterials. 2009;30:2015–2022. doi: 10.1016/j.biomaterials.2008.12.081. [DOI] [PubMed] [Google Scholar]
- 31.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glud SZ, Sørensen AB, Andrulis M, Wang B, Kondo E, Jessen R, Krenacs L, Stelkovics E, Wabl M, Serfling E, et al. A tumor-suppressor function for NFATc3 in T-cell lymphomagenesis by murine leukemia virus. Blood. 2005;106:3546–3552. doi: 10.1182/blood-2005-02-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lum AM, Wang BB, Li L, Channa N, Bartha G, Wabl M. Retroviral activation of the mir-106a microRNA cistron in T lymphoma. Retrovirology. 2007;4:5. doi: 10.1186/1742-4690-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 35.Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brudno M, Chapman M, Gottgens B, Batzoglou S, Morgenstern B. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics. 2003;4:66. doi: 10.1186/1471-2105-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 38.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 39.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 40.Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, van Lohuizen M. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Genet. 2002;32:160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 41.Hansen GM, Skapura D, Justice MJ. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res. 2000;10:237–243. doi: 10.1101/gr.10.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J. Virol. 2003;77:2056–2062. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Sorensen AB, Wang B, Wabl M, Nielsen AL, Pedersen FS. Identification of novel Bach2 transcripts and protein isoforms through tagging analysis of retroviral integrations in B-cell lymphomas. BMC Mol. Biol. 2009;10:2. doi: 10.1186/1471-2199-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 46.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, et al. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J, Du J, Rozowsky J, Zhang Z, Urban A, Euskirchen G, Weissman S, Gerstein M, Snyder M. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 2008;9:R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J-i, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trinklein ND, Karaoz U, Wu J, Halees A, Force Aldred S, Collins PJ, Zheng D, Zhang ZD, Gerstein MB, Snyder M, et al. Integrated analysis of experimental data sets reveals many novel promoters in 1% of the human genome. Genome Res. 2007;17:720–731. doi: 10.1101/gr.5716607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bajic VB, Tan SL, Christoffels A, Schonbach C, Lipovich L, Yang L, Hofmann O, Kruger A, Hide W, Kai C, et al. Mice and men: their promoter properties. PLoS Genet. 2006;2:e54. doi: 10.1371/journal.pgen.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagasaki H, Arita M, Nishizawa T, Suwa M, Gotoh O. Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene. 2005;364:53–62. doi: 10.1016/j.gene.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 58.Amoui M, Baylink DJ, Tillman JB, Lau KH. Expression of a structurally unique osteoclastic protein-tyrosine phosphatase is driven by an alternative intronic, cell type-specific promoter. J. Biol. Chem. 2003;278:44273–44280. doi: 10.1074/jbc.M303933200. [DOI] [PubMed] [Google Scholar]
- 59.Rigault C, Le Borgne F, Demarquoy J. Genomic structure, alternative maturation and tissue expression of the human BBOX1 gene. Biochim. Biophys. Acta. 2006;1761:1469–1481. doi: 10.1016/j.bbalip.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 60.Streb JW, Kitchen CM, Gelman IH, Miano JM. Multiple promoters direct expression of three AKAP12 isoforms with distinct subcellular and tissue distribution profiles. J. Biol. Chem. 2004;279:56014–56023. doi: 10.1074/jbc.M408828200. [DOI] [PubMed] [Google Scholar]
- 61.Turner JD, Schote AB, Macedo JA, Pelascini LP, Muller CP. Tissue-specific glucocorticoid receptor expression, a role for alternative first exon usage? Biochem. Pharmacol. 2006;72:1529–1537. doi: 10.1016/j.bcp.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Walsh NC, Cahill M, Carninci P, Kawai J, Okazaki Y, Hayashizaki Y, Hume DA, Cassady AI. Multiple tissue-specific promoters control expression of the murine tartrate-resistant acid phosphatase gene. Gene. 2003;307:111–123. doi: 10.1016/s0378-1119(03)00449-9. [DOI] [PubMed] [Google Scholar]
- 63.Zhang T, Haws P, Wu Q. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res. 2004;14:79–89. doi: 10.1101/gr.1225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen BP, Liang G, Whelan J, Hai T. ATF3 and ATF3 delta Zip. Transcriptional repression versus activation by alternatively spliced isoforms. J. Biol. Chem. 1994;269:15819–15826. [PubMed] [Google Scholar]
- 66.Preiss T, Hentze MW. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 67.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 69.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 70.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 73.Weidenfeld-Baranboim K, Hasin T, Darlyuk I, Heinrich R, Elhanani O, Pan J, Yokoyama KK, Aronheim A. The ubiquitously expressed bZIP inhibitor, JDP2, suppresses the transcription of its homologue immediate early gene counterpart, ATF3. Nucleic Acids Res. 2009;37:2194–2203. doi: 10.1093/nar/gkp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J. Biol. Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 75.Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J. 2005;24:2425–2435. doi: 10.1038/sj.emboj.7600712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.