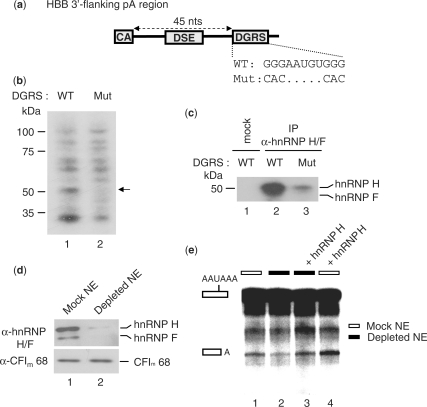
Figure 4.
HnRNP H stimulates 3′ end cleavage at the HBB pA signal by binding to a downstream GRS element. (a) Illustration of the HBB region downstream of the cleavage site (CA) and including a GRS located 45 nucleotides downstream of the cleavage site (DGRS). The sequence difference between the wild-type (WT) and mutated DGRS (Mut) is shown. (b) UV crosslinking of NE proteins to the uniformly 32P-labeled r17/HBB pA signal RNA substrate containing the WT or Mut DGRS sequence. The 50-kDa protein crosslinked to the WT but not to the Mut RNA substrate is indicated by the arrow. (c) IP of UV-crosslinked complexes described in panel (b) with the hnRNP H/F antibody followed by SDS–PAGE analysis. (d) NEs depleted of endogenous hnRNP H/F without (mock) or with (depleted) a biotinylated RNA oligonucleotide corresponding to the SVL GRS. NEs were analyzed by SDS–PAGE followed by western blot analysis. (e) In vitro cleavage assays using the 32P-labeled r17/HBB substrate in the presence of mock-depleted NEs (white boxes) or NEs depleted of hnRNP H/F (black boxes), supplemented or not with recombinant hnRNP H (1 pmol). The low cleavage efficiency is due to the depletion procedure. Identities of the uncleaved and cleaved products are shown on the left.