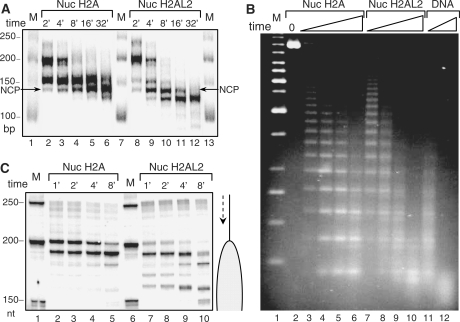
Figure 3.
Higher accessibility of H2AL2 mono-nucleosomes and nucleosomal arrays to micrococcal nuclease and exonuclease III digestion. (A) Identical amounts (50 ng) of conventional (Nuc H2A) and H2AL2 (Nuc H2AL2) nucleosomes (reconstituted on a body-labeled 255 bp 601 sequence) in a solution of 10 μl were digested (in the presence of 1 μg plasmid DNA) with 8 units/ml of micrococcal nuclease for the indicated times (2–32 min) at room temperature. After arresting the reaction, the digested DNA was isolated and run on 10% native PAGE. Lanes 1, 7 and 13, DNA molecular mass markers. The lengths (in bp) of the markers are indicated at the left side of the figure. (B) Micrococcal nuclease digestion of conventional H2A and histone variant H2AL2 33X200–601 arrays. Hundred nanogram of fully saturated reconstituted conventional (lanes 2–5), H2AL2 (lanes 7–10) and naked DNA (lanes 11 and 12) arrays were digested for different time points with micrococcal nuclease. The digested DNA was isolated and run on 1.4% agarose gel and visualized with SYBR green. Lane 1, 1-kb molecular mass DNA marker. (C) Exonuclease III digestion of conventional and H2AL2 nucleosomes. Fifty nanogram of uniquely 5′-end-labeled centrally positioned nucleosomes (reconstituted on a 255 bp 601 sequence) were digested with the same amount of exonuclease III for the times indicated. The reaction was arrested and, after purification, the digestion products were run on an 8% denaturing gel. The lengths of the 50-bp DNA marker (lanes 1 and 6) are indicated at the left side of the figure.