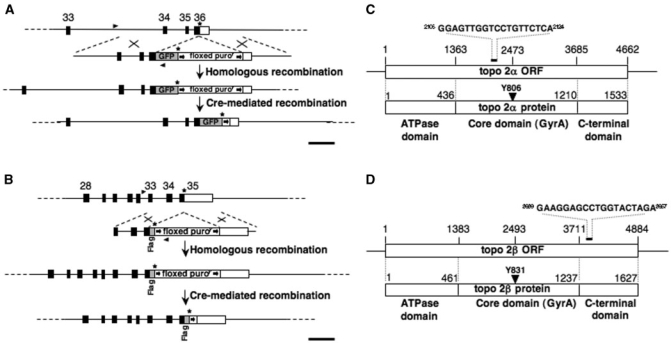
Figure 2.
(A) Map showing the 3′ region of the Gallus gallus topo 2β genomic locus and the GFP in situ tagging (IST) construct. A homologous recombination event will result in exon 36, which contains the stop codon, being disrupted, such that the portion encoding the final part of the 2β open reading frame is fused in-frame to sequence encoding eGFP, followed by a stop codon, a floxed puromycin-resistance gene cassette and sequence from the 3′ homology block. (B) Map showing the 3′ region of the chicken topo 2α genomic locus and the Flag IST construct. In the event of gene targeting, exon 35, which contains the stop codon, will be disrupted such that the protein-encoding portion of the exon is fused in-frame to a Flag epitope. This is immediately followed by a stop codon, the floxed puroR marker and sequence from the 3′ homology block. In each case, homologously recombined clones were initially identified by PCR of genomic DNA amplified using forward primers (arrowheads) lying 5′ to the homology blocks and with reverse primers based on the GFP or puroR genes. In targeted-tagged clones, the floxed puroR gene was removed by transient expression of Cre recombinase. Black boxes represent the position of exons. Exon numbering is based on the Gallus gallus reference sequence assembly 2.1. Scale bar, 1 kb. (C) Schematic showing the Gallus gallus topo 2α mRNA and its open reading frame (ORF), the various domains of the protein and the position of the RNAi target sequence (5). (D) Schematic showing the Gallus gallus topo 2β mRNA and ORF, the various domains of the protein and the position of the RNAi target sequence (5).