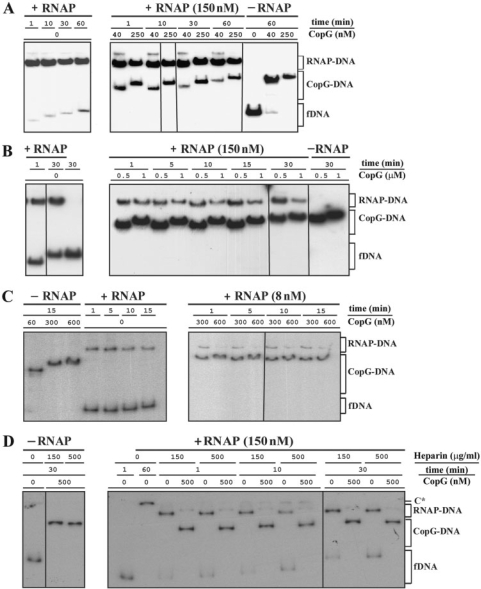
Figure 6.
CopG-mediated rapid dissociation of the RNAP–Pcr complexes at 37°C. (A, B, C, D) EMSA from four independent time-course experiments performed over a range of CopG and RNAP concentrations. RNAP holoenzyme was purchased from Boehringer (panels A, B and C) or from USB (panel D). DNA (2 nM) and RNAP at the indicated concentrations were equilibrated at 37°C and then treated at t = 0 with heparin (150 μg/ml in A and B, 10 μg/ml in C, and either 150 μg/ml or 500 μg/ml in D) followed by different concentrations of CopG. Incubation of the mixtures continued for several times before loading onto the gel. CopG–DNA complexes formed in the absence of RNAP were also analyzed. Bands corresponding to free DNA (fDNA) and to RNAP–DNA and CopG–DNA complexes are indicated. At high CopG concentrations (>200 nM), CopG–DNA complexes migrating slower than the specific complex generated by binding of four repressor dimers were observed. All the lanes shown in each experiment came either from the same gel or from gels prepared and run in parallel. Note that in the absence of heparin, slowly migrating RNAP–DNA complexes that contained several RNAP molecules were observed (C* in panel D). The percentages of RNAP–DNA and CopG–DNA complexes quantified from the EMSA displayed in panels A, B, C and D, are shown in Supplementary Tables 1, 2, 3 and 4, respectively.