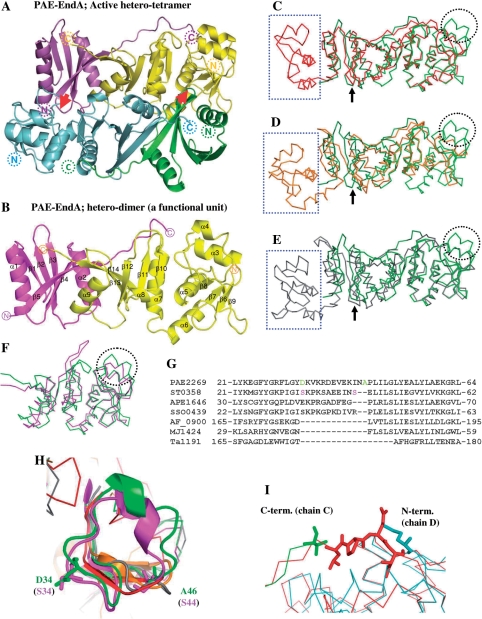
Figure 1.
Crystal structure of PAE-EndA. (A) Schematic representation of a crystal structure of PAE-EndA. The unit is composed of two catalytic subunits (cyan and yellow) and two structural subunits (green and magenta). Red arrows indicate L10-loop-like interactions between the structural and the catalytic subunits to form a hetero-tetramer. The positions of the N-termini and C-termini of the proteins are shown as dotted circles. (B) A functional hetero-dimer unit of PAE-EndA (chains A and B). The positions of the N-termini and C-termini of the proteins are shown as circles. (C–E) Superimposed structure of the PAE-EndA (PDB ID: 2ZYZ, chains A and B) on; (C) AFU-EndA (PDB ID: 1RLV, chain A); (D) TAC-EndA (PDB ID: 2OHC, chain A); and (E) MJA-EndA (PDB ID: 1A78, chains A and B). (F) Catalytic subunits of crenarchaeal origin, STO-EndA (PDB ID: 2CV8, chain B), and PAE-EndA (PDB ID: 2ZYZ, chain D) were superimposed. Through (C) to (F), color-assignments are as follows: dark green for PAE-EndA structural subunit; light green for PAE-EndA catalytic subunit; red for AFU-EndA; orange for TAC-EndA; pink for STO-EndA; gray for MJA-EndA. Positions for L10-like loop are indicated with black arrows. The EndA N-subdomain missing in the PAE-EndA structural subunit is enclosed by blue-dotted square. The extra loop missing in the euryarchaeal EndA catalytic subunits is circled with black dots. (G) Part of an amino acid sequence alignment for EndA catalytic domains or subunits. Amino acid sequences from catalytic subunits of crenarchaeal origin [PAE2269 (P. aerophilum, residues 1–183. GenBank accession number AAL64075), ST0358 (S. tokodaii, residues 1–180. Accession number BAB65337), APE1646 (Aeropyrum pernix, residues 1–187. Accession number BAA80647), and SSO0439 (S. solfataricus, residues 1–182. Accession number AAK40764)], catalytic domains of euryarchaeal homodimeric EndAs, [AF_0900 (A. fulgidus, residues 150–305. Accession number AAB90338), and Ta1191 (T. aciophilum, residues 156–289. Accession number CAC12316)], and a subunit of a euryarchaeal homotetrameric EndA, MJ1424 (M. jannaschii, residues 1–179. Accession number ABW02570) were aligned with CLUSTALW version 1.83 (43) on DNA Data Bank Japan (DDBJ) server (http://clustalw.ddbj.nig.ac.jp/top-j.html) with default parameters. (H) Magnified view of crenarchaeal EndA-specific extra loop. Residues for D34 and A46 of PAE-EndA (green) and S34 and S44 of STO-EndA catalytic subunits are presented as sticks. Color assignments for the structures are the same as in Figure 1C–F. (I) Magnified view of the superposed structure around the linker-loop region. Green, C-terminal region of the PAE-EndA structural subunit; cyan, N-terminal region of the PAE-EndA catalytic subunit; red, interdomain loop region of AF-EndA. The C-terminal residue of the PAE-EndA structural subunit (valine), the N-terminal residue of the PAE-EndA catalytic subunit (methionine), and the reference residues (-LPEI-) in the loop region for linker variants are shown as sticks.