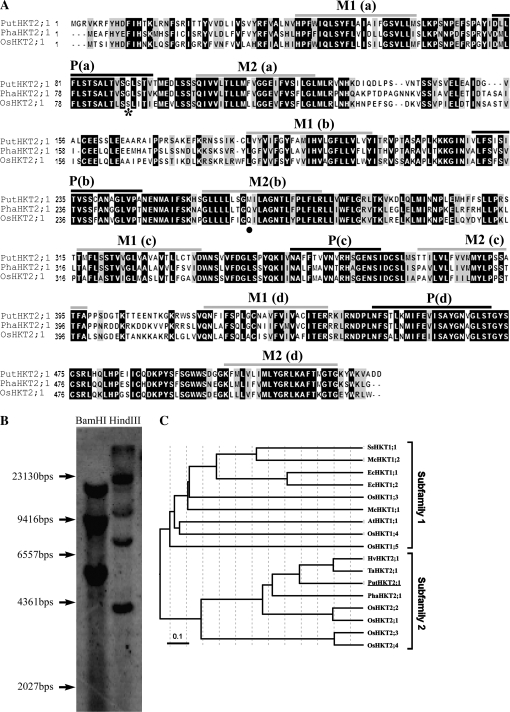
Fig. 2.
(A) Amino acid sequence alignment of PutHKT2;1 with PhaHKT2;1 (Phragmites australis, BAE44385) and OsHKT2;1 (Oryza sativa, BAB61789). The sequences were aligned by the program ClustalW. Black and grey backgrounds indicate identical residues and similar residues, respectively. Grey and black bars above the sequences indicate the putative transmembrane (M) and pore (P) domains, respectively, by the TMpred program. The conserved Gly residue is indicated by an asterisk, while the Gln270 of TaHKT2;1 which is conserved in PhaHKT2;1 and OsHKT2;1 is indicated by a black dot. Putative M1PM2 motifs (a–d) are marked. (B) Genomic Southern hybridization of PutHKT2;1. Genomic DNA (20 μg) was digested by BamHI and HindIII. Hybridization was performed overnight at 45 °C, with a digoxigenin (DIG)-labelled PutHKT2;1 cDNA probe. (C) Unrooted minimum-evolution tree of the HKT transporters. The translated sequences of known HKT genes were aligned by the ClustalW program and the tree was constructed using the MEGA 4 program (Kumar et al., 2008). Scale bar indicates 0.1 substitutions per site.