Abstract
In addition to drought and extreme temperatures, soil salinity represents a growing threat to crop productivity. Among the cereal crops, barley is considered as notably salt tolerant, and cultivars show considerable variation for tolerance towards salinity stress. In order to unravel the molecular mechanisms underlying salt stress tolerance and to utilize the natural genetic variation of barley accessions, a series of hydroponics-based salinity stress experiments was conducted using two genetic mapping parents, cvs Steptoe and Morex, which display contrasting levels of salinity tolerance. The proteome of roots from both genotypes was investigated as displayed by two-dimensional gel electrophoresis, and comparisons were made between plants grown under non-saline and saline conditions. Multivariate analysis of the resulting protein patterns revealed cultivar-specific and salt stress-responsive protein expression. Mass spectrometry-based identification was successful for 26 out of 39 selected protein spots. Hierarchical clustering was applied to detect similar protein expression patterns. Among those, two proteins involved in the glutathione-based detoxification of reactive oxygen species (ROS) were more abundant in the tolerant genotype, while proteins involved in iron uptake were expressed at a higher level in the sensitive one. This study emphasizes the role of proteins involved in ROS detoxification during salinity stress, and identified potential candidates for increasing salt tolerance in barley.
Keywords: Barley genotypes, mass spectrometry, proteome analysis, salinity stress
Introduction
Crop plants are exposed to various biotic and abiotic stress factors under field conditions, and yield reduction caused by these stresses can reach up to 50% (Vij and Tyagi, 2007). Salinity is one of the most severe abiotic stress factors, and today >800 million ha of land are salt affected, an area equivalent to 6% of the world's total land (Munns, 2005). With global food production having to meet the demands of a growing world population, improving salt tolerance of crops is an important global priority, and has become the focus for ongoing breeding efforts.
Salt tolerance is a multigenic trait, including salt partitioning within the plant, osmotic adjustment, and morphological changes (Munns, 2005). Numerous studies using the genetic model plant Arabidopsis thaliana (L.) have been aimed at the dissection of salinity tolerance (Moller and Tester, 2007). Valuable information on cellular and subcellular Na+ transport has been revealed by the dissection of the salt overly sensitive pathway (Shi et al., 2000), and the identification of a cyclic nucleotide-gated channel (Guo et al., 2008). Components of signal transduction pathways have been identified, e.g. a transmembrane protein with an ankyrin-repeat motif influencing the abscisic acid-induced accumulation of ROS (Sakamoto et al., 2008), and nuclear small ubiquitin-like modifier proteases involved in protein modification processes under salinity stress (Conti et al., 2008). In crop plants, extensive efforts have been devoted to the generation of salt-tolerant genotypes, and conventional breeding has moved towards the exploitation of transgenics, large-scale transcript and protein profiling data in conjunction with quantitative trait locus (QTL) identification (Sahi et al., 2006).
Improvements in proteomic technology regarding protein separation and detection, as well as mass spectrometry-based protein identification, have an increasing impact on the study of plant responses to salinity stress (Parker et al., 2006; Qureshi et al., 2007; Caruso et al., 2008). New insights have been obtained into salinity stress responses by comparative proteome studies of salt-stressed roots from Arabidopsis and rice (Oryza sativa). The identification of novel protein candidates associated with salinity stress (Yan et al., 2005; Jiang et al., 2007), detection of alterations in protein phosphorylation patterns (Chitteti and Peng, 2007), and, recently, the location of a salinity stress-responsive protein to the rice root apoplast with a putative function in stress signalling (Zhang et al., 2009) indicate the importance of ion uptake and transport, and regulation of water status and signal transduction processes in the root.
Mapping populations represent a valuable natural genetic resource that can be exploited for the discovery of genes and gene products conferring tolerance to various stresses. Barley (Hordeum vulgare L.) is one of the most salt-tolerant crops, and the parents of the Steptoe/Morex mapping population, a population developed to accumulate favourable agronomic traits (Hayes et al., 1996), respond differentially in terms of germination under saline conditions (unpublished data). Although the ability of a crop to germinate under unfavourable environmental conditions such as saline soil is a crucial trait, it is not predictive of its potential to produce biomass or set seed under these conditions. Therefore, the present study aimed at the assessment of salt stress responses at later developmental stages using long-term hydroponic stress experiments with the contrasting barley genotypes Steptoe and Morex. The response of barley roots challenged with salinity stress has been widely assessed at the transcript level, though not as yet at the gene product level (Ozturk et al., 2002; Ueda et al., 2002, 2006; Walia et al., 2006). Furthermore, only little attention has been paid to the genetic variability for salt tolerance in barley as shown for rice and wheat (Moons et al., 1995; Wang et al., 2008). Thus, this study set out to compare the root proteomes of two contrasting barley cultivars by 2-D gel electrophoresis. The intention was to identify genotype- and treatment-specific alterations in the protein complement, and to exploit these as potential candidate proteins involved in conferring salinity tolerance in barley.
Materials and methods
Plant material and hydroponic stress treatment
Plant culture and stress treatments were carried out according to Walia et al. (2006) with some modifications. Grains of cvs Steptoe and Morex were rinsed and placed on filter paper dampened with 0.1% Previcur N (Bayer CropScience, Langenfeld, Germany) for 5–7 d at 4 °C to break dormancy. Afterwards grains were transferred to a regime of 16 h light at 22 °C, 8 h dark at 20 °C for 2 d. Germinated seedlings were planted in a synthetic medium (Biolaston PVC) soaked in half-strength modified Hoagland's solution (Hoagland and Arnon, 1950) consisting of 3 mM KH2PO4, 1 mM MgSO4, 1 mM CaCl2·2H2O, 25 μM H3BO3, 2 μM MnSO4, 2 μM ZnSO4, 0.5 μM CuSO4, 0.5 μM Na2MoO4·2H2O, 0.1 mM Fe-EDTA, 1 mM H2SO4 and 8 mM NH4NO3 with the pH adjusted to 7.0 using KOH under 16 h light at 22 °C and 8 h dark at 20 °C for 3 d. Thereafter the seedlings were acclimated for a further 2 d in full-strength modified Hoagland's solution under aerated hydroponics. The hydroponic system was covered with a perforated plate that held the plants and conditions were adjusted to give a light intensity of 350 μEinstein, 18 °C/16 °C for 14 h/10 h light/dark, respectively, 70% humidity, with constant aeration to the nutrient solution. For reasons of stable supply with nutrients, the solutions were exchanged every other day. The salinity treatment started with the addition of NaCl to make a 50 mM solution in all tanks, except those holding the control plants. After 2 d, 100 mM NaCl was added to all tanks, except for the control and 50 mM NaCl-treated plants. The salt concentration in the nutrient solution was increased up to 250 mM following this scheme, to give salinity treatments of 0, 50, 100, 150, 200, and 250 mM NaCl. Plants were harvested after 13 d of stress application either for protein extraction or for the determination of growth parameters. All experiments were performed in triplicate.
Plant growth measurements
Measurements were taken of the dry weight of the second and third leaf and the roots of 20 individual plants of both cultivars grown at 0, 50, 100, 150, 200, and 250 mM NaCl in three independent experiments. Relative growth inhibition caused by salt stress was determined from the ratio between performance under control and stressed conditions. Statistical differences were analysed following the Duncan multiple range tests (Duncan, 1955).
Protein extraction
The extraction of proteins from roots was performed followed the trichloroacetic acid (TCA)/acetone precipitation method (Amme et al., 2005). Briefly, the frozen root material was homogenized under liquid nitrogen to a fine powder. One part (∼1 g) of this material was mixed with 10 parts of TCA/acetone solution [10% (w/v) TCA, 0.07% (w/v) 2-mercaptoethanol in acetone] and incubated for 45 min at –20 °C. The precipitate was pelleted by centrifugation and washed twice with 0.07% (w/v) 2-mercaptoethanol in acetone. The protein pellet was dried in a vacuum centrifuge and dissolved by shaking at 37 °C for 1 h in 8 M urea, 2% CHAPS, 20 mM dithiothreitol (DTT), 0.5% immobilized pH gradient (IPG) buffer. Insoluble material was removed by centrifugation at room temperature for 15 min. The 2-D Quant Kit (GE Healthcare, München, Germany) was used for the determination of protein concentration following the manufacturer's instructions.
Two-dimensional gel electrophoresis
The electrophoretic separation of the root proteome followed the methods of Schlesier and Mock (2006). Each 200 μg protein sample was loaded by rehydration to a 13 cm IPG strip using pH gradients of 3–10. The separation on an IPGphor II unit (GE Healthcare, München, Germany) was performed with the following parameters: 14 h rehydration, 1 h gradient to 250 V, 1 h gradient to 500 V, 1 h gradient to 4000 V, and 5.5 h at 4000 V with a total of ∼25 kVh. The strip was then equilibrated for 15 min in 50 mM Tris-HCl, pH 8.8, 6 M urea, 30% (v/v) glycerin, 2% (w/v) SDS, 20 mM DTT, and 0.01% bromphenol blue, placed on top of an 11.25% SDS–polyacrylamide gel, and covered with 0.5% agarose. Separation in the second dimension was performed using a Hoefer S600 (GE Healthcare, München, Germany). Afterwards, gels were washed for 5 min with water. Three independent separations of each sample were performed to ensure technical reproducibility.
Protein staining and image analysis of two-dimensional gel patterns
Proteins were visualized with GelCodeBlue Stain Reagent (Pierce Thermo Scientific, Rockford, IL, USA) following the manufacturer's instructions. Gel images were captured by a UMAX Power Look III scanner (Umax Systems, Willich, Germany) with the MagicScan software (v4.5, Umax Systems).
Delta2D software (v3.6, Decodon, Greifswald, Germany) was used for image analysis. Following the processing workflow, gel images were aligned using the ‘Group warping strategy’ and fused to an artificial gel image with the ‘Union fusion type’ function. Spots were detected under application of the default parameters. The 10 most heavily stained spots were excluded from the normalization process, since minor variation in their intensity was expected to have a large impact on the normalization of the whole data set. Statistical and visual evaluation of data was performed with tools embedded in the Delta2D software. Principle component analysis (PCA) was carried out to assess biological and technical outliers. Spot intensities between samples were compared by t-test, using the following settings: Welch approximation, P-values based on permutation of 100 randomly grouped samples, P >0.05, and false discovery control of 10 false significant data points. Hierarchical clustering was based on Euclidean distance and average linkage clustering.
Western blotting
Immunoblotting analysis was performed as described in Amme et al. (2005). The blots were probed with the monoclonal 3B6 catalase antibody from tobacco (Chen et al., 1993, kindly provided by Daniel F Klessig), a pumpkin ascorbate peroxidase antibody (Yamaguchi et al., 1995, kindly provided by Mikio Nishimura), a cucumber lipoxygenase antibody (Sturm et al., 1985, kindly provided by Helmut Kindl), and antibodies raised against the tobacco homologue of the stress-inducible protein F23N19.10 (Hedtmann and Mock, unpublished results). Signal intensity was determined using AIDA v4.19 (Raytest, Straubenhardt, Germany).
Protein identification
Spots selected were excised from 2-D gels, washed, and digested with trypsin as described in Witzel et al. (2007). The acquisition of peptide mass fingerprint data was performed on a REFLEX III MALDI-TOF (matrix-assisted laser desorption time-of-flight) mass spectrometer (Bruker Daltonics, Bremen, Germany) operating in reflector mode. Spectra were calibrated using external calibration, with subsequent internal mass correction. Protein identification was performed with the MASCOT search engine (Matrix Science, London, UK) searching the barley EST (expressed sequence tag) Gene Index in the TIGR database. Parameters for the search were as follows: monoisotopic mass accuracy 100 ppm tolerance, missed cleavages 1, allowed variable modifications: oxidation (Met) and propionamide (Cys). When this approach failed, the samples were subjected to analysis by nanoLC-ESI-Q-TOF MS/MS (nanoliquid chromatography-electrospray ionization-quadrupole time-of-flight tandem mass spectrometry) and de novo sequencing, following Amme et al. (2006). A 10 ppm peptide, 0.1 Da fragment tolerance, one missed cleavage and variable oxidation (Met) and propionamide (Cys) were used as the search parameters. Database searches were conducted against the barley EST Gene Index of the TIGR database.
Results
Evaluation of plant growth responses to salinity stress
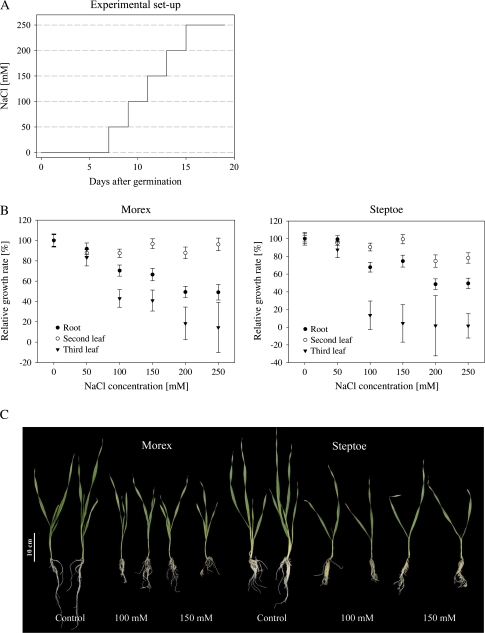
The contrasting salinity stress response of cvs Steptoe and Morex was verified by the imposition of salinity stress at the seedling stage. For long-term experiments, the hydroponic culture system was used as it facilitates the controlled application of NaCl to plants via a nutrient solution and the harvest of roots as compared with soil-grown plants. In order to determine the appropriate NaCl concentration for the subsequent proteomic analysis, plants were exposed to gradual salt stress as indicated in Fig. 1A. At the harvest time point both lines suffered a delay in growth due to the imposition of salinity stress, and both root dry weight and the dry weight of the leaves were reduced (Fig. 1B and Supplementary Table S1 available at JXB online). The growth was inhibited in both cultivars, although even at the higher salt concentrations there was no sign of any leaf senescence. No change in the biomass production of the second leaf was detected for Morex, whereas the growth of the cv. Steptoe second leaf was reduced at the highest concentrations of NaCl. The development of the cv. Steptoe third leaf was compromised at 150 mM NaCl and almost completely inhibited at 200 mM NaCl. In contrast, the cv. Morex was able to develop its third leaf even at 250 mM NaCl, although there was a growth reduction of ∼90% as compared with control plants. The root biomass was also affected by salt treatment, but there was no cultivar-specific response, as the biomass was reduced by 40–50% at the highest salt concentrations tested as compared with control plants. Based on these data, the diagnostic concentrations for the proteome analysis were set at 100 mM and 150 mM NaCl, as these generate a moderate stress response in both genotypes (Fig. 1C).
Fig. 1.
The effect of NaCl on the seedling growth of cvs Morex and Steptoe. (A) Schematic diagram of salt stress application to barley seedlings using hydroponic culture. NaCl treatment started 7 d after germination in a step-wise manner until the desired concentration was reached. Plants were harvested after 13 d of stress treatment at the indicated concentrations. (B) The relative growth rate of salinity-stressed barley seedlings. Growth inhibition in both barley cultivars was already detectable at 50 mM NaCl treatment and increased with higher salt concentrations. The biomass production of the second and third leaf in cv. Morex was more advanced than in cv. Steptoe. The data represent the means of 20 plants per treatment, with the standard error shown as error bars. (C) The effect of 100 mM and 150 mM NaCl on seedling growth.
Comparative proteome analysis
Preliminary experiments revealed only minor changes in the leaf proteome, whereas the expression of a number of root proteins was modified by salinity (data not shown). Based on these findings, an in-depth proteome analysis of root tissue from cvs Steptoe and Morex was conducted to analyse proteins in response to salt stress.
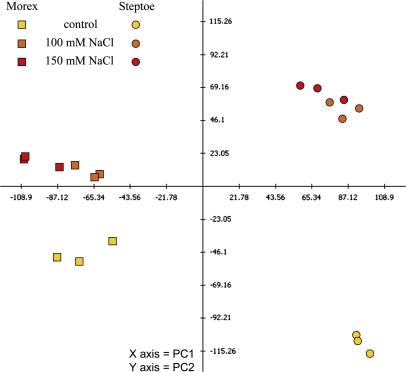
2-D gels from three biological experiments were analysed to detect proteins that were reproducibly regulated in the same manner either cultivar specifically or in response to the stress treatment. Approximately 1100 protein spots were detected on the 2-D gels, of which ∼760 were matched between groups of technical replicates. PCA indicated a grouping of technical replicates and a distinct separation of genotypes as well as treatments (Fig. 2). The contrast between salinity-stressed and non-stressed samples identified salinity-induced alterations in the protein profile. Based on the greater difference between control and treated profiles in cv. Steptoe as compared with cv. Morex, it is concluded that more proteins are affected by stress-specific regulation in the less tolerant genotype.
Fig. 2.
Assessment of technical and biological variation in protein expression profiles of control and treated samples from cvs Steptoe and Morex. Used for calculation were differentially regulated spots with P <0.05. Principle component (PC) 1 revealed genotype-specific expression and accounted for 38.6% of the variation, while PC 2 showed treatment-responsive expression and accounted for 20.9% of the variation.
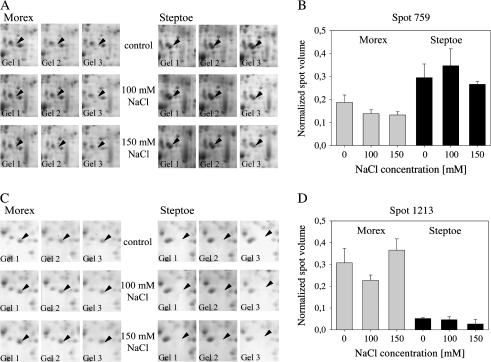
The comparative image analysis allowed for 39 differentially expressed spots to be detected based on P <0.05. Here, most spots showed a cultivar- rather than treatment-specific expression, and two examples are given in Fig. 3. The appearance on the 2-D gel and the calculated normalized volume of spot 759 revealed that this spot was more highly abundant in the root proteome of Steptoe than Morex. The expression of this spot in Steptoe was not significantly affected by the treatment, whereas a decrease in spot abundance was detected in Morex upon salinity. Spot 1213 was more highly expressed in Morex, and was down-regulated by salt stress application in cv. Steptoe.
Fig. 3.
Protein expression of spots 759 and 1213 in cv. Steptoe and cv. Morex under non-saline and saline conditions. (A, C) Section of 2-D gel for close-up view of the expression of spots 759 and 1213, respectively. (B, D) Protein spot abundance given by the normalized spot volume as determined by the image analysis software.
Identification of differentially expressed proteins
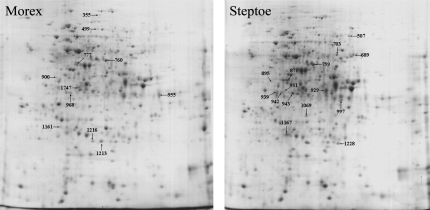
Protein spots detected as differentially expressed were excised manually from 2-D gels for tryptic digest and subjected to mass spectrometry. In order to verify their identity, protein spots were analysed from all genotypes, treatments, and two biological experiments. When the identification could not be confirmed under all conditions or failed completely due to the limited number of entries for barley in protein or EST databases, the respective protein spot was rejected from the data set. Hence, mass spectrometry analysis allowed the identity of 26 out of the 39 differentially expressed proteins. The gel migration points of these are shown in Fig. 4 and the results of the comparative proteome analysis are presented in Table 1. Detailed information relating to peptide mass fingerprinting and de novo sequencing data are provided in Supplementary Table S2 at JXB online.
Fig. 4.
Coomassie-stained 2-D gels from root samples of cvs Morex (left) and Steptoe (right) show the position of the spots listed in Table 1. The spot is indicated in the genotype where its expression was highest.
Table 1.
Root proteins differentially expressed under control and salinity stress conditions in the tolerant cv. Morex and sensitive cv. Steptoe genotype
| Spot ID | TIGR | Description | Functional category | Theoretical MW (kDa)/pI | Morex |
Steptoe |
||||
| Control | 100 mM NaCl | 150 mM NaCl | Control | 100 mM NaCl | 150 mM NaCl | |||||
| Expression not affected by salt stress | ||||||||||
| 895 | TC139604 | Late embryogenesis abundant protein, Oryza sativa | Desiccation tolerance | 41.07/4.98 | 0.023 | 0.023 | 0.019 | 0.050 | 0.041 | 0.060 |
| 900 | TC139604 | Late embryogenesis abundant protein, Oryza sativa | Desiccation tolerance | 41.07/4.98 | 0.085 | 0.094 | 0.086 | 0.040 | 0.057 | 0.045 |
| 968 | TC130772 | Lactoylglutathione lyase, Oryza sativa | Redox regulation | 32.55/5.51 | 0.217 | 0.202 | 0.223 | 0.068 | 0.067 | 0.065 |
| Up-regulated in response to salt stress in at least one genotype | ||||||||||
| 777 | TC131046 | S-Adenosylmethionine synthetase 1, Hordeum vulgare | Primary metabolism | 42.84/5.49 | 0.259 | 0.299 | 0.294 | 0.114 | 0.122 | 0.135 |
| 1161 | TC139656 | Carboxymethylenebutenolidase-like protein, Oryza sativa | Redox regulation | 30.41/6.31 | 0.145 | 0.160 | 0.200 | 0.073 | 0.088 | 0.097 |
| 955 | TC140370 | Peroxidase, Hordeum vulgare | Redox regulation | 36.55/5.91 | 0.830 | 0.957 | 0.878 | 0.537 | 0.570 | 0.559 |
| 355 | TC146955 | Lipoxygenase 1, Hordeum vulgare | Primary metabolism | 96.39/5.73 | 0.062 | 0.054 | 0.052 | 0.017 | 0.024 | 0.037 |
| 997 | TC149802 | (1–3)-β-Glucanase GV, Hordeum vulgare | Disease/defence | 34.41/6.91 | 0.265 | 0.283 | 0.256 | 0.278 | 0.354 | 0.381 |
| Down-regulated in response to salt stress in at least one genotype | ||||||||||
| 703 | TC133105 | Cytosolic 6-phosphogluconate dehydrogenase, Oryza sativa | Primary metabolism | 51.58/6.58 | 0.093 | 0.072 | 0.088 | 0.157 | 0.092 | 0.094 |
| 939 | NP315772 | Probable nicotianamine synthase 7, Hordeum vulgare | Secondary metabolism | 35.24/5.10 | 0.440 | 0.464 | 0.377 | 0.592 | 0.374 | 0.468 |
| 1747 | TC137024 | Not found | 0.266 | 0.232 | 0.207 | 0.195 | 0.150 | 0.132 | ||
| 499 | TC139384 | F23N19.10 stress-inducible protein, Arabidopsis thaliana | Disease/defence | 67.32/6.24 | 0.050 | 0.039 | 0.024 | 0.011 | 0.024 | 0.013 |
| 759 | TC132873 | Putative monodehydroascorbate reductase, Oryza sativa | Redox regulation | 52.75/6.84 | 0.187 | 0.160 | 0.139 | 0.288 | 0.307 | 0.256 |
| 760 | 35_16328 | Putative nuclear RNA-binding protein A, Oryza sativa | Protein synthesis | 40.42/6.37 | 0.388 | 0.311 | 0.236 | 0.095 | 0.077 | 0.083 |
| 931 | TC130772 | Lactoylglutathione lyase, Oryza sativa | Redox regulation | 32.55/5.51 | 0.050 | 0.022 | 0.037 | 0.203 | 0.219 | 0.200 |
| 507 | TC139323 | Poly(A)-binding protein, Triticum aestivum | Protein synthesis | 70.82/6.60 | 0.149 | 0.151 | 0.130 | 0.162 | 0.034 | 0.060 |
| 689 | TC139229 | Catalase 1, Hordeum vulgare | Redox regulation | 56.58/6.68 | 0.189 | 0.161 | 0.180 | 0.291 | 0.163 | 0.189 |
| 871 | TC137786 | Iron deficiency-specific protein IDS2, Hordeum vulgare | Secondary metabolism | 37.57/5.17 | 0.158 | 0.165 | 0.160 | 0.381 | 0.317 | 0.338 |
| 929 | TC142112 | Iron deficiency-specific protein IDS3, Hordeum vulgare | Secondary metabolism | 37.85/5.81 | 0.609 | 0.658 | 0.612 | 1.491 | 1.073 | 0.968 |
| 942 | TC147014 | Fructokinase 2, Oryza sativa | Primary metabolism | 35.51/5.02 | 0.071 | 0.074 | 0.061 | 0.236 | 0.202 | 0.209 |
| 943 | TC147167 | Iron deficiency-induced protein IDI2, Hordeum vulgare | Protein synthesis | 38.57/5.44 | 0.151 | 0.130 | 0.127 | 0.371 | 0.313 | 0.300 |
| 1167 | TC145151 | Iron deficiency-induced protein IDI1, Hordeum vulgare | Secondary metabolism | 23.46/5.23 | 0.562 | 0.451 | 0.478 | 0.915 | 0.662 | 0.736 |
| 1213 | TC146774 | Glutathione S-transferase F5, Triticum aestivum | Redox regulation | 23.43/5.78 | 0.316 | 0.323 | 0.330 | 0.076 | 0.040 | 0.053 |
| 1228 | TC138639 | 23 kDa jasmonate-induced protein, Hordeum vulgare | Disease/defence | 22.84/5.92 | 0.092 | 0.087 | 0.097 | 0.254 | 0.178 | 0.119 |
| Opposite regulation within genotypes | ||||||||||
| 1069 | TC131931 | Probable L-ascorbate peroxidase 7, Oryza sativa | Redox regulation | 38.32/8.76 | 0.073 | 0.057 | 0.042 | 0.196 | 0.290 | 0.298 |
| 1216 | TC131931 | Probable L-ascorbate peroxidase 7, Oryza sativa | Redox regulation | 38.32/8.76 | 0.256 | 0.198 | 0.153 | 0.460 | 0.443 | 0.512 |
TIGR and HarvEST databases identifiers, along with molecular weight (MW) and isoelectric point (pI) as calculated with ExPASy tools are shown. The expression level of each protein spot is indicated by the mean percentage volume of three biologically independent experiments. Statistical differences between genotypes and treatments were analysed using a paired t-test. Significant differences with P <0.05 between genotypes are underlined, and within genotype treatment-specific differences are highlighted in bold.
Twenty-four protein spots showed a cultivar-dependent expression in the absence of salinity stress, with a total of 10 spots more abundant in roots of the tolerant Morex line, and 14 spots more highly expressed in the sensitive Steptoe line. Overall, the expression of 23 protein spots was affected by salinity treatment, while the abundance of three spots was not affected by the treatment. Based on the expression profiles under salt stress conditions, the proteins were grouped into four categories. Proteins in the first category followed an expression pattern characterized by a genotype-specific expression, which was not significantly affected by salt stress treatment. Among those were two spots identified as late embryogenesis abundant protein (spots 895 and 900) and a lactoylglutathione lyase (also known as glyoxalase I). Proteins in the second category were significantly more highly expressed under stress conditions in at least one genotype. Those were lipoxygenase 1, S-adenosylmethionine synthase 1, peroxidase, (1–3)-β-glucanase, and a carboxymethylenebutenolidase-like protein. The largest group contained proteins that were down-regulated in at least one genotype upon stress treatment, and these were 6-phosphogluconate dehydrogenase, a probable nicotianamine synthase 7, F23N19.10 (a stress-inducible protein), a putative monodehydroascorbate reductase (MDAR), a putative nuclear RNA-binding protein A, lactoylglutathione lyase, a poly(A)-binding protein, catalase 1, the iron deficiency-specific proteins IDS2 and IDS3, fructokinase 2, the iron deficiency-induced proteins IDI1 and IDI2, glutathione S-transferase (GST) F5, and a 23 kDa jasmonate-induced protein. Spot 1747 was the product of barley EST TC137024, a gene lacking any significant homologue in other plant species. The last category comprises spots that were regulated in an opposite manner between genotypes and, here, one protein was found in two distinct spots (spots 1069 and 1216), identified as a probable L-ascorbate peroxidase 7.
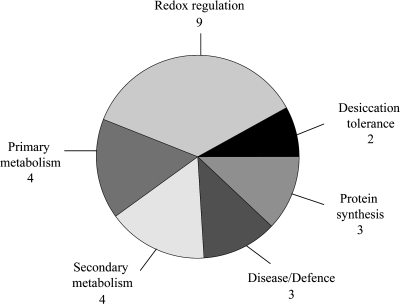
According to their functional annotation, most protein spots identified in the analysis are part of the oxidative stress responses, with eight protein spots functioning in redox regulation. The remaining proteins are involved in protein synthesis, primary and secondary metabolism, desiccation tolerance, and disease/defence-related processes (Fig. 5).
Fig. 5.
Functional classification of proteins detected in the comparative proteome analysis.
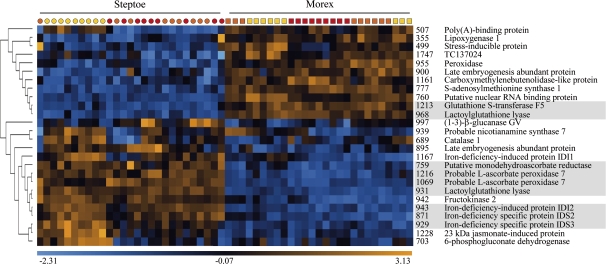
Hierarchical clustering of genotype-specific and salt stress-responsive proteins
In order to identify proteins with similar expression patterns, a hierarchical clustering method was applied. Two main clusters of protein expression were found, with the first consisting of proteins that were more abundant in cv. Morex and the second containing proteins that were more highly expressed in cv. Steptoe (Fig. 6). Within these clusters it was possible to recognize three groups of proteins sharing similar biological function (Fig. 6, highlighted in grey). The first group consisted of (GST) F5 (spot 1213) and lactoylglutathione lyase (spot 968), both involved in redox regulation. The latter was more abundant in cv. Morex than in cv. Steptoe, and its level of expression was not induced by salinity. GST F5 expression was significantly altered only at 100 mM NaCl in cv. Steptoe. Members of the second group were also related to the protein redox state and included a putative MDAR (spot 759), two spots identified as a putative L-ascorbate peroxidase 7 (spots 1069 and 1216), and lactoylglutathione lyase (spot 931). Those proteins were down-regulated upon salinity treatment in cv. Morex but not in cv. Steptoe. The expression of both spots of putative L-ascorbate peroxidase 7 was even induced in Steptoe upon treatment, indicating genotype- and treatment-specific regulation. The third group comprised three proteins involved in secondary metabolism: the iron deficiency-induced protein IDI2 (spot 943) and the iron deficiency-specific proteins IDS2 (spot 871) and IDS3 (spot 929), all of which were significantly down-regulated only in the sensitive Steptoe genotype.
Fig. 6.
Hierarchical clustering of expression patterns of differentially regulated proteins. Each column represents one 2-D gel prepared from samples of 0 mM NaCl- (yellow), 100 mM NaCl- (orange) and 150 mM NaCl- (red) treated plants of genotypes Steptoe (circles) and Morex (squares) in three biological replicates per treatment. Rows display the abundance of the protein spot on 2-D gels using colour coding based on a relative scale (–2.31 to +3.13), as derived from normalized spot volumes. Spot numbers and protein names are listed for each row. Groups of proteins with similar biological function are highlighted in grey.
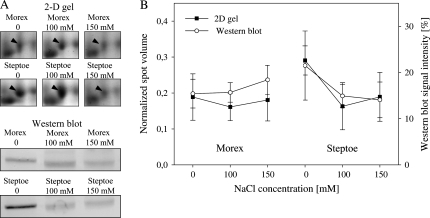
Validation of spot expression by immunoblotting analysis
In order to confirm the protein expression revealed by 2-D gel electrophoresis, western blot analysis was performed on a selected number of proteins. Available antibodies against catalase, lipoxygenase, ascorbate peroxidase, and the stress-inducible protein F23N19.10 were used for immunoblot experiments. Overall, a high degree of commonality in protein expression was found when normalized spot volumes were compared with signals obtained from immunoblotting. An example is given in Fig. 7 for catalase (spot 689). Shown are the normalized spot volumes determined by the image analysis software and the signal intensity of the western band assessed by densitometric measurement. Here, a higher expression was detected by both methods in cv. Steptoe under control conditions. Upon salinity treatment, a reduction in protein abundance was detected by both approaches in cv. Steptoe, whereas in cv. Morex no significant change was observed. Similar tendencies between spot expression and western signal were retrieved using the remaining antisera (data not shown).
Fig. 7.
Estimated protein abundance of catalase (689) based on 2-D gel electrophoresis and western blot analysis. (A) Protein expression in one biological experiment using 2-D gel electrophoresis and immunoblotting. (B) Normalized spot volume as revealed by 2-D gels, and western blot signal intensity determined by densitometric measurement of western blot bands. Error bars show the standard deviation of measurements based on three independent biological experiments.
Discussion
Assessment of salinity tolerance at the seedling stage of cv. Steptoe and cv. Morex
The salt stress experiments conducted here using the contrasting genotypes Steptoe and Morex aimed at comparative root proteome analysis in order to identify candidate proteins for salinity tolerance in barley. Despite the large body of physiological studies on salt stress in various plant species, most of the underlying molecular mechanisms are still unclear.
For assessing growth responses of Steptoe and Morex towards salinity, hydroponic stress experiments were conducted. Both cultivars responded to salinity stress by an inhibition of plant growth, but, in contrast to the leaves, where growth reduction of up to 90% was noticed, root growth was less severely compromised (40–50%). The reduction of water uptake is an early outcome of salinity stress and there are different mechanisms controlling growth at different time points for plants that are exposed to salinity. There are rapid changes in leaf and root growth within a few minutes to hours after stress application, but this fast response is mainly due to the changes in cell water relations, and not to salt stress (Munns, 2002). A few days after the imposition of salinity treatment, the leaf and root growth is stably reduced, with leaf elongation more inhibited than root growth. This phenomenon can also be observed for plants subjected to water deficiency (Hsiao and Xu, 2000), and evidence indicates that abscisic acid signalling is primarily responsible for this reaction.
Proteomic analysis of salt-stressed root tissue
Roots are exposed directly to salt and control ion uptake from the soil as well as transport within the plant. Comparative proteomic studies of salt-stressed roots of Arabidopsis and rice have allowed new insights into the plant response to salinity stress, such as novel proteins associated with stress responses and the regulation of post-translational modifications (Yan et al., 2005; Chitteti and Peng, 2007; Jiang et al., 2007). For both model plants vast molecular genetic resources are available, but nevertheless most genotypes of these two species are sensitive to only moderate levels of salinity. In contrast to this, barley is considered as notably salt tolerant among the cereals, and available accessions cover a wide range of responses towards salinity. Barley germplasm analysis has consistently identified the presence of ample genetic variation for salinity tolerance, and this has been exploited to study large-scale gene expression in roots of cvs Haruna-nijyo, Tokak, and Morex under salt stress (Ozturk et al., 2002; Ueda et al., 2002, 2006; Walia et al., 2006). However, information on the expression of gene products is still scarce. In the present study, the analysis of the proteome is targeted and MS-based identification of genotype- or treatment-specific proteins was successful for 26 spots. The number of differentially regulated proteins is low in comparison with other studies. Proteome analysis of Arabidopsis roots stressed with 150 mM NaCl revealed the regulation of ∼200 protein spots (Jiang et al., 2007), and ∼110 proteins were changed in expression in contrasting wheat genotypes exposed to 200 mM NaCl (Wang et al., 2008). However, those species are relatively sensitive towards salinity (Munns and Tester, 2008) and the salt levels chosen are likely to provoke unspecific stress reactions rather than leading to the discovery of gene products specifically related to salt tolerance. In order to avoid provoking osmotic shock of plants, a step-wise addition of NaCl was chosen, as described for cv. Morex (Walia et al., 2006), and moderate levels of NaCl (100 mM and 150 mM) were applied to trigger cultivar-specific stress responses.
Correlations of protein expression patterns revealed by hierarchical clustering
Comparative proteome analysis led to the detection of proteins involved in a variety of biological processes, such as protein synthesis, redox regulation, primary and secondary metabolism, or disease- and defence-related processes. Hierarchical clustering of protein expression profiles was applied and resulted in two major groups of proteins that were more abundant in cv. Morex or in cv. Steptoe under control and stress conditions. With the intention of detecting functionally related proteins that have a similar regulation pattern, three groups involved in redox and in secondary metabolism processes were found. Proteins in the first group were more abundant in the tolerant line cv. Morex and were identified as GST and lactoylglutathione lyase, also known as glyoxalase I. The generation and scavenging of ROS is tightly regulated in plants, with ∼150 genes involved in these networks in Arabidopsis (Miller et al., 2008). This indicates a high level of specificity as well as redundancy in gene function since ROS play a role in numerous biological processes, e.g. biotic and abiotic stress responses, growth, development, and hormone signalling (Mittler et al., 2004). GST catalyses the conjugation of reduced glutathione with a variety of target compounds, and its involvement in oxidative stress responses is known (Droog, 1997); its overexpression in tobacco has been associated with enhanced salinity tolerance during germination (Roxas et al., 1997). Lactoylglutathione lyase is involved in the glutathione-based detoxification of methylglyoxal (MG), a toxic byproduct of carbohydrate and amino acid metabolism. MG is formed both by the fragmentation and elimination of phosphate from the phospho-ene-diolate form of glyceraldehyde-3-phosphate and dihydroxyacetone phosphate, and from the oxidation of aminoacetone during threonine biosynthesis (Thornalley, 1996). The accumulation of MG is indicative of abiotic stress conditions, such as salinity, drought, and cold (Yadav et al., 2005a). The toxic effect of MG ranges from mutagenization of nucleic acids to modification with subsequent degradation of proteins (Thornalley, 1996). Two enzymes are involved in the detoxification of MG. Glyoxalase I catalyses the conversion to S-D-lactoylglutathione and glyoxalase II catalyses the hydrolysis to D-lactate under the release of glutathione. Recently it was shown that tobacco plants overexpressing glyoxalase I possessed a higher salinity tolerance than wild-type plants (Singla-Pareek et al., 2003), an effect which was accentuated in plants co-transformed with glyoxalase I and II. Plants overexpressing both glyoxalase enzymes were able to maintain a higher reduced to oxidized glutathione ratio under salinity stress (Yadav et al., 2005b). The detoxification of ROS and the maintenance of the protein redox balance are crucial mechanisms in salt stress tolerance. The constitutively higher expression of proteins involved in glutathione-based ROS scavenging in the more tolerant cv. Morex could represent a pre-formed tolerance mechanism.
Proteins forming the second group in the hierarchical tree are also related to redox regulation. The proteins whose level of expression is higher in cv. Steptoe than in cv. Morex include MDAR, two spots of different molecular weight identified as L-ascorbate peroxidase (APX), and lactoylglutathione lyase, which was identified in a second spot with a higher molecular weight as compared with the protein spot in the first group. The induction of APX by salt stress treatment was not unexpected, since it catalyses the reduction of H2O2 to water with the concomitant generation of monodehydroascorbate, and is therefore a key component in the scavenging pathway of ROS produced by various stress factors (Shigeoka et al., 2002). The accumulation of APX upon salinity treatment is in agreement with the literature (Shigeoka et al., 2002; Jiang et al., 2007; Caruso et al., 2008; Wang et al., 2008), but, surprisingly, the initial APX level was greater in the sensitive genotype Steptoe than in the tolerant Morex. Also more abundant in cv. Steptoe was MDAR, which catalyses the regeneration of ascorbate (Mittler et al., 2004). In a proteomic analysis of Arabidopsis roots subjected to salt stress, MDAR expression was inhibited, leading to the proposal that although the requirement for reduced ascorbate is high under salinity stress, the fine-tuning of antioxidant levels is also important (Jiang et al., 2007). Lactoylglutathione lyase was identified in a protein spot closely located to the respective spot in group 1, but showed a contrasting expression as compared with the former. The observation of multiple spots with different expression under salt stress conditions could point to a regulation mechanism based on multiple isoforms or post-translational modifications.
Proteins in the third group were involved in iron uptake (IDI2, IDS2, and IDS3) and were expressed initially at a higher level in cv. Steptoe compared with cv. Morex. Their expression declined under salinity stress in the former, but was not changed in the latter. IDI2 was initially isolated as cDNA from Fe-deficient barley roots and encodes a protein related to the α-subunit of the eukaryotic translation initiation factor 2B, probably regulating the synthesis of proteins required for stress adaptation (Yamaguchi et al., 2000). IDS2 and IDS3 are two dioxygenase genes isolated from iron-deficient barley roots (Nakanishi et al., 2000). Iron-deficient graminaceous plants produce phytosiderophores which are secreted into the rhizosphere and facilitate iron uptake (Curie and Briat, 2003). Phytosiderophores have a high affinity for iron and solubilize Fe3+ by chelation; the resulting complexes are then transported through the plasma membrane via specific transporters. The substrate of IDS2 and IDS3 is 2′-deoxymugineic acid, which is metabolized to synthesize mugineic acid family phytosiderophores. The reduced protein expression of IDI2, IDS2, and IDS3 indicates a salinity stress-induced growth inhibition and a concomitant decrease in iron consumption, but it may also reflect the avoidance of metal ion-induced oxidative stress. Although iron is an essential cofactor for many proteins, its reaction with oxygen can lead to the production of hydroxyl and peroxide radicals in the Fenton reaction. These radicals are engaged in secondary reactions such as protein oxidation, lipid peroxidation, and DNA nicking (Briat, 2002). Limiting iron uptake under salt stress conditions could therefore counteract ROS formation.
Candidate proteins for augmenting salinity tolerance in barley
A number of cultivar-specific and salt stress-responsive proteins have been identified here, and some of these represent potential candidates for augmenting salt tolerance in barley. Possible candidates for further in-depth analysis could be proteins induced upon stress treatment in the tolerant cultivar or proteins down-regulated in the sensitive cultivar. Also, proteins that were more abundant in Morex and where the expression was not affected by the salt treatment could be of interest. The expression of S-adenosylmethionine (SAM) synthase was induced by salinity stress in both genotypes, but protein expression was higher in cv. Morex than in cv. Steptoe. This enzyme catalyses the attachment of an adenosyl residue to methionine, resulting in the formation of SAM, which is a universal methyl group donor for DNA, proteins, carbohydrates, membrane lipids, flavonoids, and others (Roje, 2006). It is also the precursor of ethylene, polyamides, nicotianamine, phytosiderophores, and biotin (Roje, 2006). Proteomic analysis of Arabidopsis germinating and developing seedlings has shown that SAM synthase is strongly accumulated (Gallardo et al., 2001), and that germination can be inhibited by blocking methionine synthesis (Gallardo et al., 2002). An enhanced methionine metabolism could therefore promote plant development under abiotic stress conditions.
One of the induced proteins which was more abundant in cv. Morex was a carboxymethylenebutenolidase-like protein featuring a dienelactone hydrolase (DLH) domain. DHL domains are responsible for the detoxification of chloroaromatic compounds in bacteria (Bruckmann et al., 1998). In a proteomic study of the rice aleurone layer, DLH was identified as a putative target for thioredoxin-mediated reduction (Yano and Kuroda, 2006). Thioredoxins reduce disulphide bonds and donate electrons to various enzymes, such as peroxiredoxins functioning in H2O2 detoxification (Dietz et al., 2006). As yet, the substrate of DLH in plants in unknown, but its induction upon salt stress treatment suggests a regulatory role in redox metabolism.
Peroxidase, a key player in ROS scavenging, was more highly expressed in cv. Morex compared with cv. Steptoe, and was induced under salt stress conditions. The encoding gene was initially cloned from barley coleoptiles, and expression was induced after infection with powdery mildew fungus (Kristensen et al., 1999). The predicted subcellular localization (http://wolfpsort.org/) places it as a class III peroxidase secreted to the cell wall or surrounding medium. Upon wounding, pathogen attack, or other unfavourable environmental conditions, ROS are supplied by an oxidative burst. Peroxidases catalyse the reduction of H2O2 by using various molecules at the cell wall as a substrate, such as phenolic compounds, lignin precursors, auxin, or others, leading to polymerization reactions such as lignification, suberization, and cross-linking of cell wall proteins (Kristensen et al., 1999; Passardi et al., 2004). The higher abundance of peroxidase and its inducibility in cv. Morex suggest that cell wall modifications may be occurring to reduce ion influx under saline conditions.
Furthermore, proteins involved in ROS scavenging, such as GST and lactoylglutathione lyase, may also be relevant for functional analysis.
Conclusions
Salinity stress is a complex trait, and tolerance mechanisms of natural genotypes are probably rather diverse. By comparing the root proteomes of a salt-tolerant and a salt-sensitive genotype, 39 proteins showing a cultivar-specific or a stress-related expression were detected, and identification was successful for 26 proteins. Candidates conferring salt tolerance include peroxidase, GST, lactoylglutathione lyase, SAM synthase, and a carboxymethylenebutenolidase-like protein. Some of these have now been selected for more detailed functional analysis. Further validation of possible candidates also includes the investigation of additional accessions from the Steptoe/Morex population that show an even stronger contrasting phenotypic performance under salinity stress as compared with the parentage.
Supplementary data
Supplementary data are available at JXB online.
Acknowledgments
The financial support to H-PM by the BMBF (GABI-SEED II, FKZ 0313115) is gratefully acknowledged. G-KS acknowledges the award of a DAAD/Leibniz post-doctoral scholarship (A/03/27383), and KW the award of a COST short-term scientific mission fellowship (FA0603-03178). Daniel F Klessig, Mikio Nishimura, and Helmut Kindl are all thanked for providing antibodies. We acknowledge the photographic work of Heike Ernst and the excellent technical assistance of Christa Kallas and Petra Linow.
Glossary
Abbreviations
- APX
ascorbate peroxidase
- GST
glutathione S-transferase
- MDAR
monodehydroascorbate reductase
- MG
methylglyoxal
- PCA
principle component analysis
- QTL
quantitative trait locus
- ROS
reactive oxygen species
References
- Amme S, Matros A, Schlesier B, Mock H-P. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. Journal of Experimental Botany. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- Amme S, Rutten T, Melzer M, Sonsmann G, Vissers JPC, Schlesier B, Mock H-P. A proteome approach defines protective functions of tobacco leaf trichomes. Proteomics. 2005;5:2508–2518. doi: 10.1002/pmic.200401274. [DOI] [PubMed] [Google Scholar]
- Briat J-F. Metal ion-activated oxidative stress and its control. In: Inze D, van Montagu M, editors. Oxidative stress in plants. London: Taylor & Francis Inc; 2002. [Google Scholar]
- Bruckmann M, Blasco R, Timmis KN, Pieper DH. Detoxification of protoanemonin by dienelactone hydrolase. Journal of Bacteriology. 1998;180:400–402. doi: 10.1128/jb.180.2.400-402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso G, Cavaliere C, Guarino C, Gubbiotti R, Foglia P, Laganà A. Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Analytical and Bioanalytical Chemistry. 2008;391:381–390. doi: 10.1007/s00216-008-2008-x. [DOI] [PubMed] [Google Scholar]
- Chen Z, Ricigliano JW, Klessig DF. Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proceedings of the National Academy of Sciences, USA. 1993;90:9533–9537. doi: 10.1073/pnas.90.20.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitteti BR, Peng ZH. Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. Journal of Proteome Research. 2007;6:1718–1727. doi: 10.1021/pr060678z. [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. The Plant Cell. 2008;20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat J-F. Iron transport and signaling in plants. Annual Review of Plant Biology. 2003;54:183–206. doi: 10.1146/annurev.arplant.54.031902.135018. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SMN, Baier M, Finkemeier I. The function of peroxiredoxins in plant organelle redox metabolism. Journal of Experimental Botany. 2006;57:1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- Droog F. Plant glutathione S-transferases, a tale of theta and tau. Journal of Plant Growth Regulation. 1997;16:95–107. [Google Scholar]
- Duncan DB. Multiple range and multiple F tests. Biometrics. 1955;11:1–42. [Google Scholar]
- Gallardo K, Job C, Groot S, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of arabidopsis seed germination and priming. Plant Physiology. 2001;126:835–848. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D. Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiologia Plantarum. 2002;116:238–247. doi: 10.1034/j.1399-3054.2002.1160214.x. [DOI] [PubMed] [Google Scholar]
- Guo KM, Babourina O, Christopher DA, Borsics T, Rengel Z. The cyclic nucleotide-gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiologia Plantarum. 2008;134:499–507. doi: 10.1111/j.1399-3054.2008.01157.x. [DOI] [PubMed] [Google Scholar]
- Hayes P, Chen F, Kleinhofs A, Kilian A, Mather D. Barley genome mapping and its applications. In: Jauhar P, editor. Method of genome analysis in plants. Boca Raton, FL: CRC Press; 1996. pp. 229–249. [Google Scholar]
- Hoagland D, Arnon D. The water culture method for growing plants without soil. Columbia Agriculture Experimental Station Circular. 1950;347 [Google Scholar]
- Hsiao TC, Xu LK. Sensitivity of growth of roots versus leaves to water stress: biophysical analysis and relation to water transport. Journal of Experimental Botany. 2000;51:1595–1616. doi: 10.1093/jexbot/51.350.1595. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. Journal of Experimental Botany. 2007;58:3591–3607. doi: 10.1093/jxb/erm207. [DOI] [PubMed] [Google Scholar]
- Kristensen BK, Bloch H, Rasmussen SK. Barley coleoptile peroxidases. Purification, molecular cloning, and induction by pathogens. Plant Physiology. 1999;120:501–512. doi: 10.1104/pp.120.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Shulaev V, Mittler R. Reactive oxygen signaling and abiotic stress. Physiologia Plantarum. 2008;133:481–489. doi: 10.1111/j.1399-3054.2008.01090.x. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Moller IS, Tester M. Salinity tolerance of Arabidopsis: a good model for cereals? Trends in Plant Science. 2007;12:534–540. doi: 10.1016/j.tplants.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Moons A, Bauw G, Prinsen E, Vanmontagu M, Vanderstraeten D. Molecular and physiological-responses to abscisic-acid and salts in roots of salt-sensitive and salt-tolerant indica rice varieties. Plant Physiology. 1995;107:177–186. doi: 10.1104/pp.107.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant, Cell and Environment. 2002;25:239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Munns R. Genes and salt tolerance: bringing them together. New Phytologist. 2005;167:645–663. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Molecular Biology. 2000;44:199–207. doi: 10.1023/a:1006491521586. [DOI] [PubMed] [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ. Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Molecular Biology. 2002;48:551–573. doi: 10.1023/a:1014875215580. [DOI] [PubMed] [Google Scholar]
- Parker R, Flowers TJ, Moore AL, Harpham NVJ. An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. Journal of Experimental Botany. 2006;57:1109–1118. doi: 10.1093/jxb/erj134. [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends in Plant Science. 2004;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Qureshi MI, Qadir S, Zolla L. Proteomics-based dissection of stress-responsive pathways in plants. Journal of Plant Physiology. 2007;164:1239–1260. doi: 10.1016/j.jplph.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Roje S. S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry. 2006;67:1686–1698. doi: 10.1016/j.phytochem.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Roxas VP, Smith RK, Allen ER, Allen RD. Overexpression of glutathione S-transferase glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nature Biotechnology. 1997;15:988–991. doi: 10.1038/nbt1097-988. [DOI] [PubMed] [Google Scholar]
- Sahi C, Singh A, Kumar K, Blumwald E, Grover A. Salt stress response in rice: genetics, molecular biology, and comparative genomics. Functional and Integrative Genomics. 2006;6:263–284. doi: 10.1007/s10142-006-0032-5. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Matsuda O, Iba K. ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. The Plant Journal. 2008;56:411–422. doi: 10.1111/j.1365-313X.2008.03614.x. [DOI] [PubMed] [Google Scholar]
- Schlesier B, Mock H-P. Protein isolation and 2-D electrophoretic separation. In: Sanchez-Serrano J, Salinas J, editors. Arabidopsis protocols. 2nd edn. Totowa, NJ: Humana Press; 2006. pp. 381–391. [Google Scholar]
- Shi HZ, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences, USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. Regulation and function of ascorbate peroxidase isoenzymes. Journal of Experimental Botany. 2002;53:1305–1319. [PubMed] [Google Scholar]
- Singla-Pareek SL, Reddy MK, Sopory SK. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proceedings of the National Academy of Sciences, USA. 2003;100:14672–14677. doi: 10.1073/pnas.2034667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Schwennesen K, Kindl H. Isolation of proteins assembled in lipid body membranes during fat mobilization in cucumber cotyledons. European Journal of Biochemistry. 1985;150:461–468. doi: 10.1111/j.1432-1033.1985.tb09044.x. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ. Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification—a role in pathogenesis and antiproliferative chemotherapy. General Pharmacology. 1996;27:565–573. doi: 10.1016/0306-3623(95)02054-3. [DOI] [PubMed] [Google Scholar]
- Ueda A, Kathiresan A, Bennett J, Takabe T. Comparative transcriptome analyses of barley and rice under salt stress. Theoretical and Applied Genetics. 2006;112:1286–1294. doi: 10.1007/s00122-006-0231-4. [DOI] [PubMed] [Google Scholar]
- Ueda A, Shi W, Nakamura T, Takabe T. Analysis of salt-inducible genes in barley roots by differential display. Journal of Plant Research. 2002;115:119–130. doi: 10.1007/s102650200017. [DOI] [PubMed] [Google Scholar]
- Vij S, Tyagi AK. Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnology Journal. 2007;5:361–380. doi: 10.1111/j.1467-7652.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close T. Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Functional and Integrative Genomics. 2006;6:143–156. doi: 10.1007/s10142-005-0013-0. [DOI] [PubMed] [Google Scholar]
- Wang M-C, Peng Z-Y, Li C-L, Li F, Liu C, Xia G-M. Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics. 2008;8:1470–1489. doi: 10.1002/pmic.200700569. [DOI] [PubMed] [Google Scholar]
- Witzel K, Surabhi G-K, Jyothsnakumari G, Sudhakar C, Matros A, Mock H-P. Quantitative proteome analysis of barley seeds using ruthenium(II)-tris-(bathophenanthroline-disulphonate) staining. Journal of Proteome Research. 2007;6:1325–1333. doi: 10.1021/pr060528o. [DOI] [PubMed] [Google Scholar]
- Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochemical and Biophysical Research Communications. 2005a;337:61–67. doi: 10.1016/j.bbrc.2005.08.263. [DOI] [PubMed] [Google Scholar]
- Yadav SK, Singla-Pareek SL, Reddy MK, Sopory SK. Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Letters. 2005b;579:6265–6271. doi: 10.1016/j.febslet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Nakanishi H, Nishizawa NK, Mori S. Isolation and characterization of IDI2, a new Fe-deficiency-induced cDNA from barley roots, which encodes a protein related to the alpha subunit of eukaryotic initiation factor 2B. Journal of Experimental Botany. 2000;51:2001–2007. doi: 10.1093/jexbot/51.353.2001. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Mori H, Nishimura M. A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant and Cell Physiology. 1995;36:1157–1162. doi: 10.1093/oxfordjournals.pcp.a078862. [DOI] [PubMed] [Google Scholar]
- Yan S, Tang Z, Su W, Sun W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics. 2005;5:235–244. doi: 10.1002/pmic.200400853. [DOI] [PubMed] [Google Scholar]
- Yano H, Kuroda M. Disulfide proteome yields a detailed understanding of redox regulations: a model study of thioredoxin-linked reactions in seed germination. Proteomics. 2006;6:294–300. doi: 10.1002/pmic.200402033. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiology. 2009;149:916–928. doi: 10.1104/pp.108.131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.