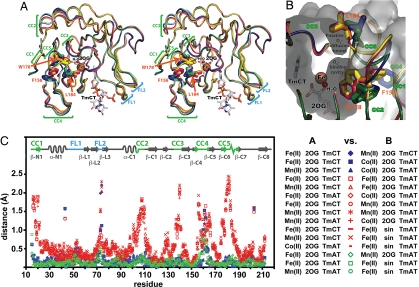
Fig. 3.
The ensemble of AlkB-ΔN11 crystal structures reveals a discrete change in global protein conformation. (A) Nucleotide-bound structures after least-squares superposition of core β-strands in the Fe-2OG dioxygenase domain [with 2OG, Fe(II), and TmCT shown from just a single structure]. Discrete local CCs are observed in 5 protein segments (CC1–CC5). FLs (FL1 and FL2), which contact the backbone of the trinucleotide substrate, exhibit elevated backbone B-factors as well as conformational variability not correlated with the conformational state of the enzyme (16). The water molecule replaced by O2 to initiate the oxidation reaction occupies the sixth site on the octahedrally coordinated Fe ion. AlkB-Fe(II)-2OG-TmCT is shown in orange, AlkB-Mn(II)-2OG-TmCT in yellow, AlkB-Fe(II)-2OG-TmAT (at 1.7 Å) in green, AlkB-Mn(II)-2OG-TmAT (16) in blue, AlkB-Co(II)-2OG-TmAT (16) in magenta, and AlkB-Fe(II)-succinate-TmAT (16) in cyan. (B) The putative O2-diffusion channel and O2-binding cavity with structures colored as in A. The molecular surface of AlkB-Fe(II)-2OG-TmAT is shown in gray. (C) Plot of Cα displacements between all nucleotide-bound crystal structures after least-squares superposition of core β-strands. The legend indicates the color/symbol used for pairwise structural comparisons. Blue and green are used for comparisons between pairs of structures both in the closed or open conformational state, respectively, whereas red is used for comparisons between structures in different states. For stereo view of B, see SI Appendix, Fig. S3.