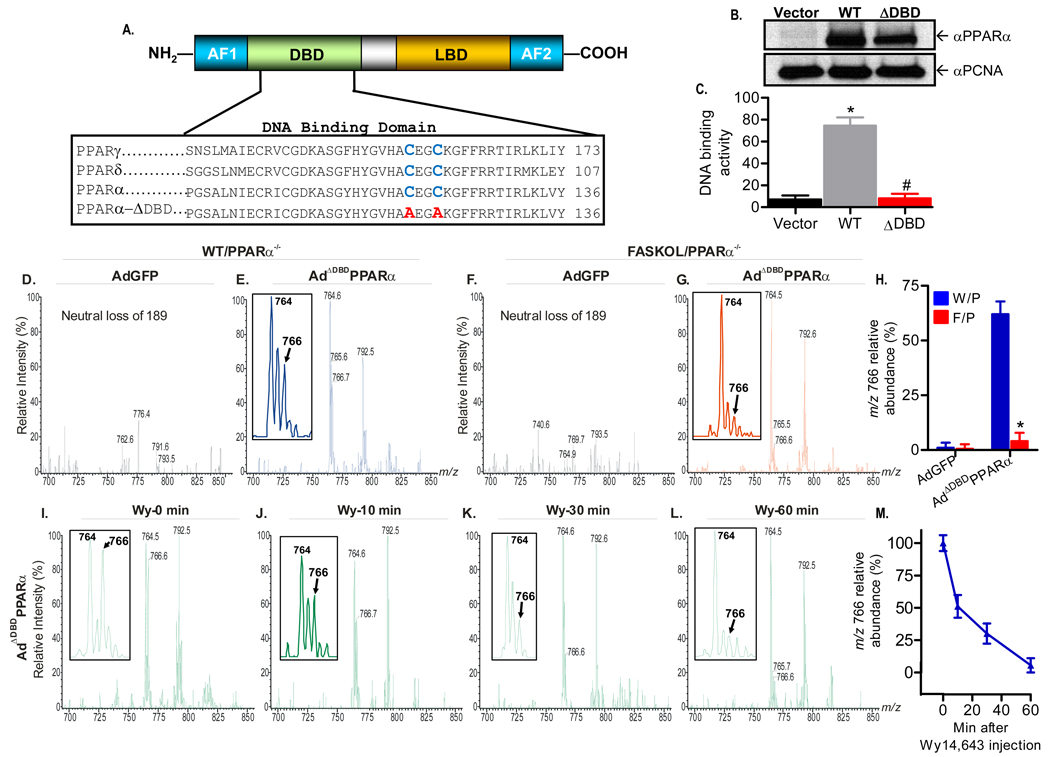
Figure 5. Generation of a PPARα DNA Binding Domain (DBD) Mutant Adenovirus and Mass Spectrometric Analysis.
(A) Schematic of the modular domain structure of PPARα (top panel). AF, activating function; LBD, ligand binding domain. The two highly conserved cysteine residues (blue) within the DBD of the PPAR family (bottom panel) were mutated to alanine (red).
(B) Immunoblot analysis of Cos-7 cells transfected with empty vector, wild type (WT), or DBD-mutant (ADBD) PPARα plasmids using anti-PPARα and proliferating cell nuclear antigen (PCNA) antibodies. Gels are representative of three independent experiments.
(C) Mutation C119A, C122A disrupts PPARα DNA binding activity. Cos-7 cells were transfected and DNA binding activity was assayed. Graphs represent mean ± SEM of experiments performed in triplicate. *, P < 0.05 vs. empty vector. #, P < 0.05 vs. WT control.
(D–G) Representative positive ion ESI/MS/MS scans monitoring neural loss of 189 from lithiated adducts of GPC species in FLAG-eluted hepatic nuclear extracts obtained from chow fed WT and FASKOL mice on a PPARα null background infected with AdGFP (D and F) or AdΔDBDPPARα
(E and G) adenoviruses. Insets in E and G indicate that the material represented by the ion at m/z 766 is the FAS-dependent phospholipid molecular species. A tandem spectrum of this ion (Figure 3) establishes its identity as [MLi+] of 16:0/18:1-GPC
(H) Quantification of the relative abundance of the m/z 766 ion in response to control and mutant adenoviral injections in W/P (WT on PPARα null background) and F/P (FASKOL on PPARα null− background) mice. Each bar represents the mean ± SEM from three independent experiments with 4–6 mice in each group per experiment. *, P < 0.05 vs. corresponding W/P control.
(I–L) Representative neutral loss of 189 ESI/MS/MS scans of GPC species at baseline (time 0) (I), 10 min (J), 30 min (K) and 60 min (L) following intraperitoneal injection of 50 µg/g Wy14,643 in chow fed WT mice on a PPARα null−/− background injected with AdΔDBDPPARα adenovirus. Insets in I–L indicate that the ion at m/z 766 (16:0/18:1-GPC) is displaced from the DBD defective PPARα in a time-dependent manner by Wy14,643.
(M) Quantification of the relative abundance of the m/z 766 ion in response to Wy14,643 administration in WT mice on a PPARα null background injected with AdΔDBDPPARα adenovirus. Graphs represent mean ± SEM from two separate experiments with 3–4 mice in each group per experiment.